9J50
 
 | |
9IR2
 
 | |
5I2O
 
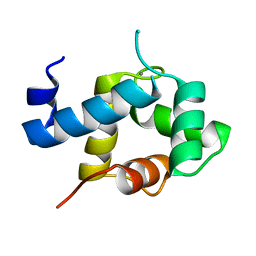 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
9JI7
 
 | | NADP-dependent oxidoreductase complexed with NADP and substrate 2 | | Descriptor: | (1Z,3E,5E,7S,8R,9S,10S,11R,13R,15S,16Z,18E,24S)-11-ethyl-2,7-dihydroxy-10-methyl-21,25-diazatetracyclo[22.2.1.08,15.09,13]heptacosa-1,3,5,16,18-pentaene-20,26,27-trione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase | | Authors: | Li, Y, Zhu, D, Xie, X, Li, F, Lu, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for Medium-Chain Dehydrogenase/Reductase-Catalyzed Reductive Cyclization in Polycyclic Tetramate Macrolactam Biosynthesis.
J.Am.Chem.Soc., 147, 2025
|
|
9JJ4
 
 | | NADP-dependent oxidoreductase complexed with NADP and substrate 1a | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase | | Authors: | Li, Y, Zhu, D, Xie, X, Li, F, Lu, M. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Medium-Chain Dehydrogenase/Reductase-Catalyzed Reductive Cyclization in Polycyclic Tetramate Macrolactam Biosynthesis.
J.Am.Chem.Soc., 147, 2025
|
|
9JM9
 
 | | NADP-dependent oxidoreductase complexed with NADP and substrate 3 | | Descriptor: | (1Z,3E,5E,7R,8S,10R,11R,12S,15S,16E,18E,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.07,15.08,12]octacosa-1,3,5,13,16,18-hexaene-20,27,28-trione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase | | Authors: | Li, Y, Zhu, D, Xie, X, Li, F, Lu, M. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural Basis for Medium-Chain Dehydrogenase/Reductase-Catalyzed Reductive Cyclization in Polycyclic Tetramate Macrolactam Biosynthesis.
J.Am.Chem.Soc., 147, 2025
|
|
9IQD
 
 | | Apo-MicM,homologous of AkaM, SnoaL-like protein | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2024-07-12 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Tandem Michael addition and Knoevenagel condensation catalyzed by NTF2-Like Enzymes as iminium catalysis
To Be Published
|
|
5NNS
 
 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
6ILD
 
 | | Crystal Structure of Human LysRS: P38/AIMP2 Complex II | | Descriptor: | 5'-O-[(S)-hydroxy(methyl)phosphoryl]adenosine, Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, GLYCEROL, ... | | Authors: | Hei, Z, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2018-10-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex.
J. Biol. Chem., 294, 2019
|
|
1AF4
 
 | | CRYSTAL STRUCTURE OF SUBTILISIN CARLSBERG IN ANHYDROUS DIOXANE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, SUBTILISIN CARLSBERG | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1997-03-21 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of subtilisin Carlsberg in anhydrous dioxane and its comparison with those in water and acetonitrile.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6S66
 
 | | Crystal structure of hTEAD2 in complex with a trisubstituted pyrazole inhibitor | | Descriptor: | 1-(2-azanylethyl)-3-(3,4-dichlorophenyl)-~{N}-(phenylmethyl)pyrazole-4-carboxamide, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Sturbaut, M, Allemand, F, Guichou, J.F. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a cryptic site at the interface 2 of TEAD - Towards a new family of YAP/TAZ-TEAD inhibitors.
Eur.J.Med.Chem., 226, 2021
|
|
6INK
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IJS
 
 | | Human PPARgamma ligand binding domain complexed with SB1494 | | Descriptor: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6S6R
 
 | |
9IVN
 
 | | Crystal structure of KRED mutant-Y199A/N149L | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Xu, H, Zhang, Z, Zhang, Y, Xu, Y, Zhang, L. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemoenzymatic synthesis of key squalamine intermediate by engineered ketoreductase mutants from Novosphingobium aromaticivorans
To Be Published
|
|
5YLG
 
 | | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A | | Descriptor: | 1,2-ETHANEDIOL, Chitinase A, ZINC ION | | Authors: | Ohnuma, T, Kitaoku, Y, Umemoto, N, Numata, T, Fukamizo, T. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A
To Be Published
|
|
5L9S
 
 | |
6I83
 
 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
6S0I
 
 | |
6S6L
 
 | | Cryo-EM structure of murine norovirus (MNV-1) | | Descriptor: | Capsid protein | | Authors: | Snowden, J.S, Hurdiss, D.L, Adeyemi, O.O, Ranson, N.A, Herod, M.R, Stonehouse, N.J. | | Deposit date: | 2019-07-03 | | Release date: | 2020-05-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM.
Plos Biol., 18, 2020
|
|
1AYU
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT SYMMETRIC BISCARBOHYDRAZIDE INHIBITOR | | Descriptor: | 1,5-BIS(N-BENZYLOXYCARBONYL-L-LEUCINYL)CARBOHYDRAZIDE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent and selective human cathepsin K inhibitors that span the active site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5KWS
 
 | | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose
To Be Published
|
|
6EIF
 
 | | DYRK1A in complex with XMD7-117 | | Descriptor: | 4-(3-pyridin-3-yl-1~{H}-pyrrolo[2,3-b]pyridin-5-yl)benzenesulfonamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5KJU
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
4U3W
 
 | |
