5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
6FFC
 
 | | Structure of an inhibitor-bound ABC transporter | | Descriptor: | ATP-binding cassette sub-family G member 2, ~{tert}-butyl 3-[(2~{S},5~{S},8~{S})-14-cyclopentyloxy-2-(2-methylpropyl)-4,7-bis(oxidanylidene)-3,6,17-triazatetracyclo[8.7.0.0^{3,8}.0^{11,16}]heptadeca-1(10),11,13,15-tetraen-5-yl]propanoate | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7QY0
 
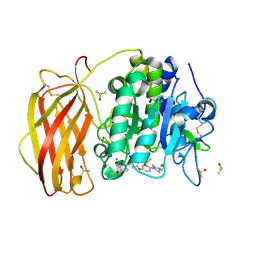 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
8S3X
 
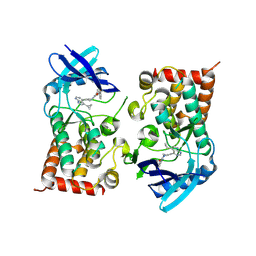 | | LIM Domain Kinase 2 (LIMK2) bound to compound 52 | | Descriptor: | 4-(5-cyclopropyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-~{N}-[3-(3-methoxyphenyl)phenyl]-3,6-dihydro-2~{H}-pyridine-1-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Ple, K, Knapp, S. | | Deposit date: | 2024-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Tetrahydropyridine LIMK inhibitors: Structure activity studies and biological characterization.
Eur.J.Med.Chem., 271, 2024
|
|
7R0M
 
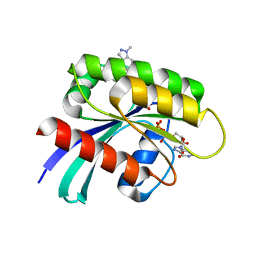 | | KRasG12C in complex with GDP and JDQ443 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-(1-methylindazol-5-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
8UF6
 
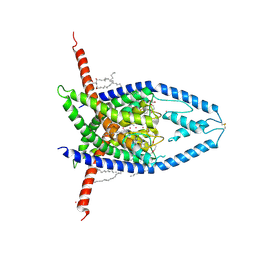 | | Structure of Trek-1(K2P2.1) with ML336 | | Descriptor: | CADMIUM ION, DECANE, DODECANE, ... | | Authors: | Lolicato, M, Mondal, A, Minor, D.L. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-26 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of covalent chemogenetic K 2P channel activators.
Cell Chem Biol, 31, 2024
|
|
8B6S
 
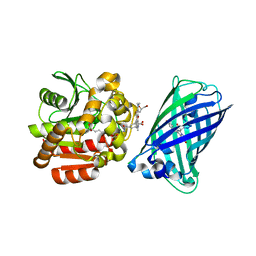 | | X-ray structure of the haloalkane dehalogenase HaloTag7 fusion to the green fluorescent protein GFP (ChemoG1) labeled with a chloroalkane tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein,Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
6REP
 
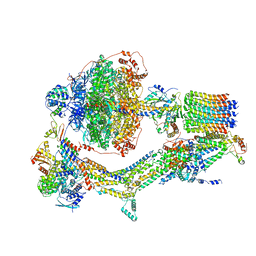 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 3, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6ZEI
 
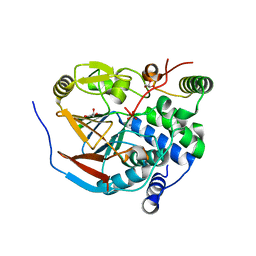 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
5HAT
 
 | | Structure function studies of R. palustris RubisCO (S59F/M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Leong, J.G, Cascio, D, Varaljay, V.A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
8HE4
 
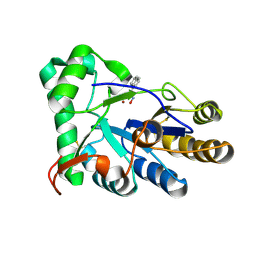 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8FC7
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK2798745 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8UGF
 
 | | In-situ complex III, state III | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
6R6K
 
 | | Structure of a FpvC mutant from pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique ferrous iron binding mode is associated with large conformational changes for the transport protein FpvC of Pseudomonas aeruginosa.
Febs J., 287, 2020
|
|
6G0H
 
 | |
6G0Q
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Erythroid transcription factor | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5HAN
 
 | | Structure function studies of R. palustris RubisCO (S59F mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Leong, J.G, Varaljay, V.A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
7BLE
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with the tricyclic inhibitor (3) | | Descriptor: | 3-ethyl-1,3-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-2-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Molecules, 26, 2021
|
|
8OJH
 
 | | Crystal structure of human CRBN-DDB1 in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-7-methoxy-isoindole-1,3-dione, DNA damage-binding protein 1, ... | | Authors: | Cabry, M.P, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
8UGG
 
 | | In-situ complex III, state IV | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8U8X
 
 | | crystal structure of the receptor tyrosine kinase Human HER2 (ERBB2) YVMA mutant kinase domain in complex with inhibitor compound 27 | | Descriptor: | 1-{(1R,3r,5S)-3-[(3M)-4-methyl-3-{3-methyl-4-[(1-methyl-1H-benzimidazol-5-yl)oxy]phenyl}-1H-pyrazolo[3,4-d]pyrimidin-1-yl]-8-azabicyclo[3.2.1]octan-8-yl}propan-1-one, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, J, Mou, T.C. | | Deposit date: | 2023-09-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Potent and Selective Covalent Inhibitors of HER2 WT and HER2 YVMA .
J.Med.Chem., 67, 2024
|
|
8OLU
 
 | |
6DO7
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6OHD
 
 | | P38 in complex with T-3220137 | | Descriptor: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
6AZN
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
