6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
7ORP
 
 | | crystal structure of human carbonic anhydrase II in complex with 4-((2-hydroxy-3-((3,4,5-trimethoxyphenyl)tellanyl)propyl)selanyl)benzenesulfonamide | | Descriptor: | 4-[(2R)-2-oxidanyl-3-(3,4,5-trimethylphenyl)tellanyl-propyl]selanylbenzenesulfonamide, 4-[(2S)-2-oxidanyl-3-(3,4,5-trimethoxyphenyl)tellanyl-propyl]selanylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Chalcogenides-incorporating carbonic anhydrase inhibitors concomitantly reverted oxaliplatin-induced neuropathy and enhanced antiproliferative action.
Eur.J.Med.Chem., 225, 2021
|
|
8DII
 
 | |
8WFE
 
 | | The Crystal Structure of PPARg from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, F, Cheng, W, Lv, Z, Guo, S, Lin, D. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of PPARg from Biortus.
To Be Published
|
|
7MLT
 
 | | Crystal structure of ricin A chain in complex with 5-(2-ethylphenyl)thiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-ethylphenyl)thiophene-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
8WFR
 
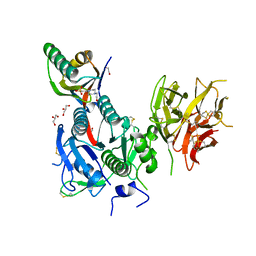 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
7RY4
 
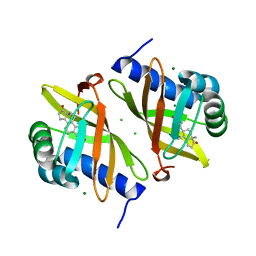 | | Multi-conformer model of Ketosteroid Isomerase Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to a transition state analog at 250 K | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-08-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ensemble-function relationships to dissect mechanisms of enzyme catalysis.
Sci Adv, 8, 2022
|
|
4RXM
 
 | | Crystal structure of periplasmic ABC transporter solute binding protein A7JW62 from Mannheimia haemolytica PHL213, Target EFI-511105, in complex with Myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of solute binding sugar transporter A7JW62 from Mannheimia haemolytica, Target EFI-511105.
To be Published
|
|
6ONP
 
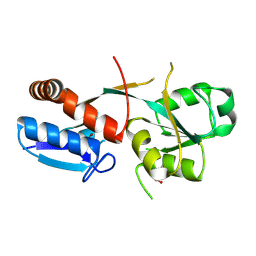 | |
7OEY
 
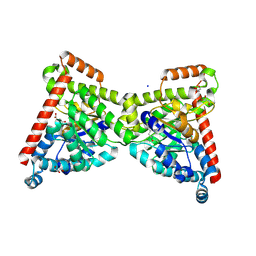 | | Neisseria gonnorhoeae variant E93Q at 1.35 angstrom resolution | | Descriptor: | GLYCEROL, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Tittmann, K. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Widespread occurrence of covalent lysine-cysteine redox switches in proteins.
Nat.Chem.Biol., 18, 2022
|
|
6R5E
 
 | |
7OJD
 
 | | SaFtsZ(D46A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7S3W
 
 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
8P9B
 
 | | Crystal Structure of Mnk2-D228G in complex with Tinodasertib | | Descriptor: | 4-[6-(4-morpholin-4-ylcarbonylphenyl)imidazo[1,2-a]pyridin-3-yl]benzenecarbonitrile, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Turnbull, A.P, Sabin, V, Bell, C, Watson, M. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of Mnk2-D228G in complex with Tinodasertib
To Be Published
|
|
7S44
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
8ZXD
 
 | | the Planar Cell Polarity Core Protein Vangl1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, Vang-like protein 1 | | Authors: | Zhang, F, Chen, S. | | Deposit date: | 2024-06-14 | | Release date: | 2024-11-27 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure and oligomerization of the human planar cell polarity core protein Vangl1.
Nat Commun, 16, 2025
|
|
6XFS
 
 | | Class C beta-lactamase from Escherichia coli in complex with Tazobactam | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Class C beta-lactamase from Escherichia coli in complex with Tazobactam
To Be Published
|
|
8WY7
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
6R11
 
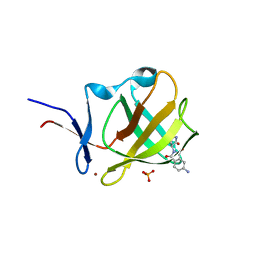 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5b | | Descriptor: | 5-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, Cereblon isoform 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
8X2S
 
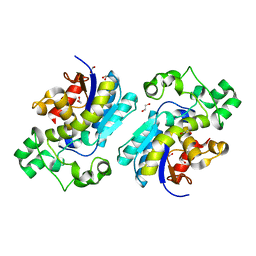 | | The Crystal Structure of BPGM from Biortus | | Descriptor: | 1,2-ETHANEDIOL, Bisphosphoglycerate mutase | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of BPGM from Biortus
To Be Published
|
|
6X7N
 
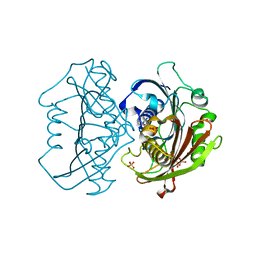 | | LnmK in complex with 2-nitronate-propionyl-amino(dethia)-CoA | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, SULFATE ION, [1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylamino]-1-oxidanylidene-propan-2-ylidene]-bis(oxidanidyl)azanium | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
9AX3
 
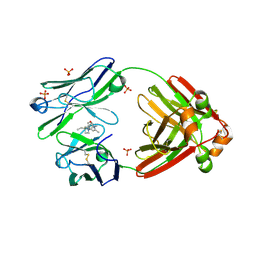 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 4-(3-ethyl-3-phenylpyrrolidin-1-yl)-1,6-dimethylpyridin-2(1H)-one | | Descriptor: | 4-[(3R)-3-ethyl-3-phenylpyrrolidin-1-yl]-1,6-dimethylpyridin-2(1H)-one, H9 Immunoglobulin Light Chain, PHOSPHATE ION | | Authors: | Lederberg, O.L, Yan, N.L, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
8XB9
 
 | | The Crystal Structure of polo-box domain of PLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase PLK1 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Meng, Q, Zhang, B. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of polo-box domain of PLK1 from Biortus
To Be Published
|
|
8X23
 
 | | The Crystal Structure of MAPK13 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mitogen-activated protein kinase 13 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-09 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of MAPK13 from Biortus.
To Be Published
|
|
7LMQ
 
 | |
