6W8X
 
 | | Cryo-EM of the S. solfataricus pilus | | Descriptor: | pilin | | Authors: | Wang, F, Baquero, D.P, Su, Z, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structures of two archaeal type IV pili illuminate evolutionary relationships.
Nat Commun, 11, 2020
|
|
6HIV
 
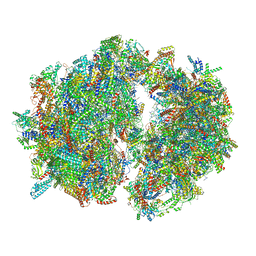 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete mitoribosome | | Descriptor: | 12S rRNA, 50S ribosomal protein L13, putative, ... | | Authors: | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boerhringer, D, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-07 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
2H8W
 
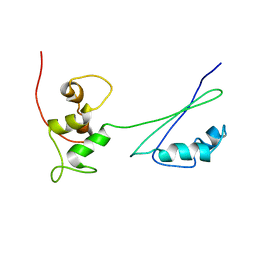 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
8DF0
 
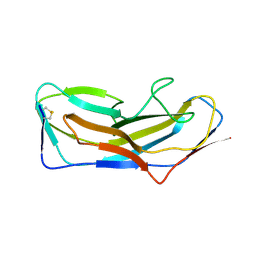 | | Abp1D receptor binding domain | | Descriptor: | Abp1D Receptor Binding Domain | | Authors: | Tamadonfar, K.O, Pinkner, J.S, Dodson, K.W, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DEZ
 
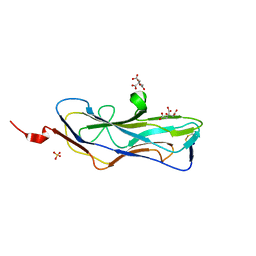 | | Abp2D Receptor Binding Domain ACICU | | Descriptor: | Abp2D Receptor Binding Domain, CITRATE ANION, SULFATE ION | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Dodson, K.W, Kalas, V, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7OEO
 
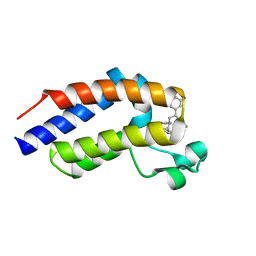 | |
7OEP
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 5-(1-(1,3-dimethoxypropan-2-yl)-5-morpholino-1H-benzo[d]imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5-[1-(1,3-dimethoxypropan-2-yl)-5-morpholin-4-yl-benzimidazol-2-yl]-1,3-dimethyl-pyridin-2-one, ... | | Authors: | Chung, C. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
7OES
 
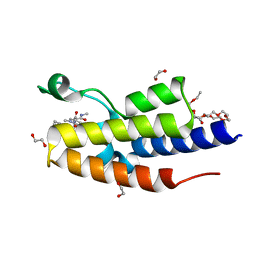 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1,5-dimethyl-N-(2-(methylamino)-2-oxoethyl)-6-oxo-N-(2-phenylpropyl)-1,6-dihydropyridine-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 1,5-dimethyl-N-[2-(methylamino)-2-oxidanylidene-ethyl]-6-oxidanylidene-N-[(2S)-2-phenylpropyl]pyridine-3-carboxamide, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
7OER
 
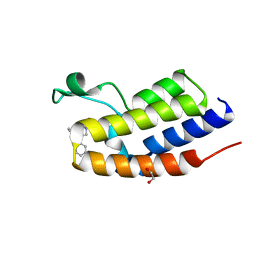 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-(2,2-diphenylethyl)-1,5-dimethyl-N-(2-(methylamino)-2-oxoethyl)-6-oxo-1,6-dihydropyridine-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-(2,2-diphenylethyl)-1,5-dimethyl-N-[2-(methylamino)-2-oxidanylidene-ethyl]-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
8EV4
 
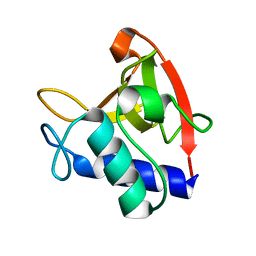 | | NlpC B3 - Trichomonas Vaginalis | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
1TFR
 
 | | RNASE H FROM BACTERIOPHAGE T4 | | Descriptor: | MAGNESIUM ION, T4 RNASE H | | Authors: | Mueser, T.C, Nossal, N.G, Hyde, C.C. | | Deposit date: | 1996-04-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of bacteriophage T4 RNase H, a 5' to 3' RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins.
Cell(Cambridge,Mass.), 85, 1996
|
|
8EV5
 
 | | NlpC B3 covalently bound with E64 inhibitor fragment | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase, ... | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
6H6A
 
 | | Crystal structure of UNC119 in complex with LCK peptide | | Descriptor: | GLY-CYS-GLY-CYS-SER-SER, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Yelland, T, ElMaghloob, Y, McIlwraith, M, Stephen, L, Ismail, S. | | Deposit date: | 2018-07-26 | | Release date: | 2018-09-26 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK.
Dev.Cell, 47, 2018
|
|
7Z5S
 
 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin, HEXAETHYLENE GLYCOL, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
7Z5T
 
 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain | | Descriptor: | ACETATE ION, Botulinum neurotoxin, PENTAETHYLENE GLYCOL | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
1TMM
 
 | | Crystal structure of ternary complex of E.coli HPPK(W89A) with MGAMPCPP and 6-Hydroxymethylpterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 6-HYDROXYMETHYLPTERIN, ACETATE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Wu, Y, Shi, G, Ji, X, Yan, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Is the Critical Role of Loop 3 of Escherichia coli 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase in Catalysis Due to Loop-3 Residues Arginine-84 and Tryptophan-89? Site-Directed Mutagenesis, Biochemical, and Crystallographic Studies.
Biochemistry, 44, 2005
|
|
6JVK
 
 | | Crystal structure of human MTH1 in complex with compound MI1012 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVT
 
 | | Crystal structure of human MTH1 in complex with compound MI1030 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-[4-(oxetan-3-yl)piperazin-1-yl]pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6EOJ
 
 | | PolyA polymerase module of the cleavage and polyadenylation factor (CPF) from Saccharomyces cerevisiae | | Descriptor: | Polyadenylation factor subunit 2,Polyadenylation factor subunit 2, Protein CFT1, ZINC ION, ... | | Authors: | Casanal, A, Kumar, A, Hill, C.H, Emsley, P, Passmore, L.A. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Architecture of eukaryotic mRNA 3'-end processing machinery.
Science, 358, 2017
|
|
6JVJ
 
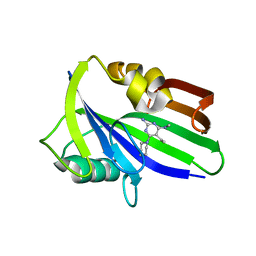 | | Crystal structure of human MTH1 in complex with compound MI1006 | | Descriptor: | 5-ethyl-N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVR
 
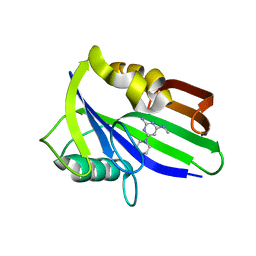 | | Crystal structure of human MTH1 in complex with compound MI1026 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
1J9K
 
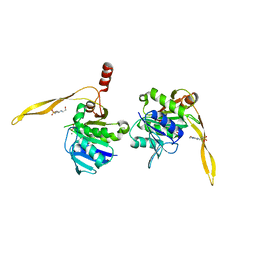 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
6JVI
 
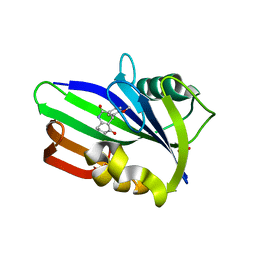 | | Crystal structure of human MTH1 in complex with compound MI0861 | | Descriptor: | (4R)-4-(2-methoxyphenyl)-4,6,7,8-tetrahydroquinoline-2,5(1H,3H)-dione, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Peng, C, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVP
 
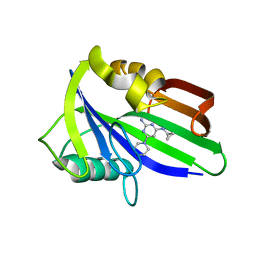 | | Crystal structure of human MTH1 in complex with compound MI1024 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
1J9L
 
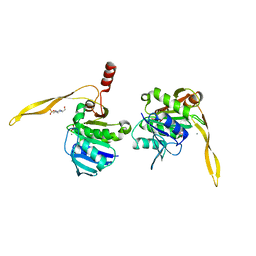 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-28 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
