8V76
 
 | |
8V6Z
 
 | |
8EKN
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 15 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({[(5P)-3-methyl-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
9QRS
 
 | | Crystal structure of CERK6 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, J.X.J, Gysel, K, Stougaard, J, Andersen, K.R. | | Deposit date: | 2025-04-04 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
8UFW
 
 | | CA9 mimic with SLC compound | | Descriptor: | 4-{[4-(trifluoromethyl)phenyl]carbamamido}benzene-1-sulfonamide, CHLORIDE ION, Carbonic anhydrase 2, ... | | Authors: | Peat, T.S. | | Deposit date: | 2023-10-04 | | Release date: | 2024-12-18 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.215 Å) | | Cite: | An ureido-substituted benzenesulfonamide carbonic anhydrase inhibitor exerts a potent antitumor effect in vitro and in vivo.
Exp Hematol Oncol, 14, 2025
|
|
9FHD
 
 | | hKHK-C in fomplex with BI-9787 | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase, SULFATE ION | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
8CFK
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment E06 | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry E06
To be published
|
|
8CFM
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment E08 | | Descriptor: | 5-(2-methoxyethyl)-1,3,4-oxadiazol-2-amine, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry E08
To be published
|
|
1EKH
 
 | | NMR STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
6FJN
 
 | | Adenovirus species 26 knob protein, 0.97A | | Descriptor: | 1,2-ETHANEDIOL, Fiber | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
9O2T
 
 | |
6V0U
 
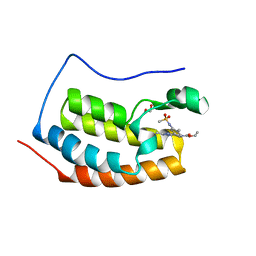 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
7PZX
 
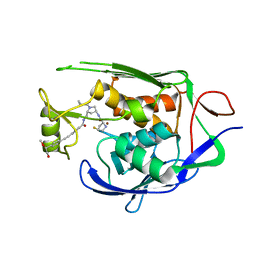 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2S,4S)-N-((3R,5R)-1-(cyclopropanecarbonyl)-5-((4-((4-((S)-2-hydroxy-1-methoxyethyl)phenyl)ethynyl)benzamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
8V5H
 
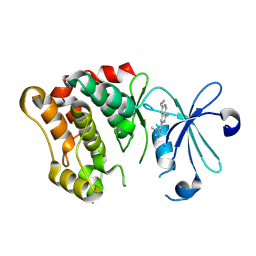 | |
6FKO
 
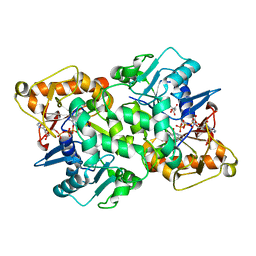 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, dGMP, hadacidin at 2.1 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, dGMP, HAdacidin at 2.1 Angstrom resolution
To Be Published
|
|
8DRE
 
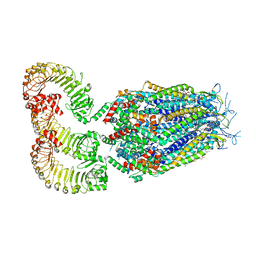 | | LRRC8A:C conformation 2 (oblong) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6MLS
 
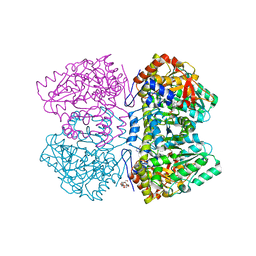 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from L-tyrosine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-09-27 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
8CVR
 
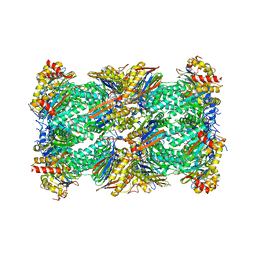 | | Human 20S proteasome with MG-132 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PRC
 
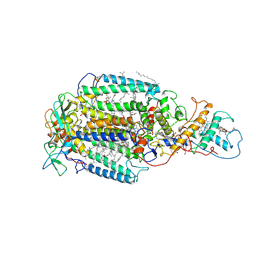 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420315 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(R(+)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-08-01 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
6V6V
 
 | |
8VDB
 
 | |
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
8VTS
 
 | | Meis1 homeobox domain bound to paromomycin fragment | | Descriptor: | 1,2-ETHANEDIOL, Homeobox protein Meis1, ISOPROPYL ALCOHOL, ... | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
6VBQ
 
 | |
8E3S
 
 | |
