7PG0
 
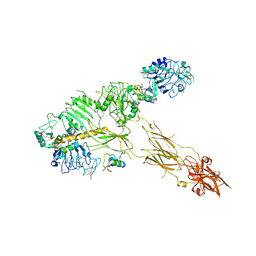 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin with visible ddm micelle, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
1FH0
 
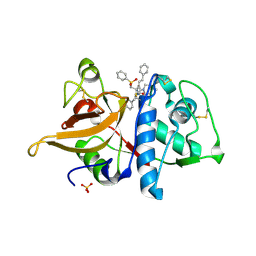 | |
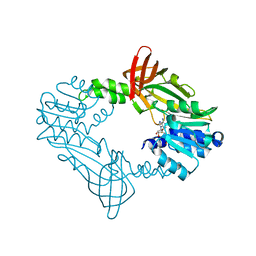
8FT7
 
 | |
8G0U
 
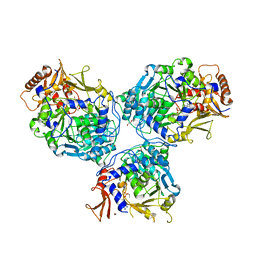 | |
8D54
 
 | |
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
6WZM
 
 | | LY3041658 Fab bound to CXCL8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-8, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
8OQK
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine kinase | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
5J63
 
 | | Crystal Structure of the N-terminal N-formyltransferase Domain (residues 1-306) of Escherichia coli Arna in Complex with UDP-Ara4N and Folinic Acid | | Descriptor: | (2R,3R,4S,5S)-5-amino-3,4-dihydroxytetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, (4aS,6S)-2-amino-6-{(E)-[(4-methylphenyl)imino]methyl}-4-oxo-4,6,7,8-tetrahydropteridine-5(4aH)-carbaldehyde, Bifunctional polymyxin resistance protein ArnA | | Authors: | Thoden, J.B, Genthe, N.A, Holden, H.M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Escherichia coli ArnA N-formyltransferase domain in complex with N(5) -formyltetrahydrofolate and UDP-Ara4N.
Protein Sci., 25, 2016
|
|
1FH9
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED LACTAM OXIME INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-(2Z,3S,4S,5R)-2-hydroxyiminopiperidine-3,4,5-triol | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
7RY5
 
 | | Cellular Retinoic Acid Binding Protein II with Bound Inhibitor 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid | | Descriptor: | 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Kimmel, B.R, Mrksich, M. | | Deposit date: | 2021-08-24 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Enzyme-Inhibitor Reaction Using Cellular Retinoic Acid Binding Protein II for One-Pot Megamolecule Assembly.
Chemistry, 27, 2021
|
|
8DBV
 
 | | E. coli ATP synthase imaged in 10mM MgATP State3 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
7RT3
 
 | | Crystal Structure of KRAS G12D with compound 24 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
6WW5
 
 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6OJ0
 
 | | Cryo-EM reconstruction of Sulfolobus polyhedral virus 1 (SPV1) | | Descriptor: | Structural protein VP4, Uncharacterized protein | | Authors: | Wang, F, Liu, Y, Conway, J.F, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A packing for A-form DNA in an icosahedral virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7P4K
 
 | | Soluble epoxide hydrolase in complex with FL217 | | Descriptor: | Bifunctional epoxide hydrolase 2, ~{N}-[[4-(cyclopropylsulfonylamino)-2-(trifluoromethyl)phenyl]methyl]-1-[(3-fluorophenyl)methyl]indole-5-carboxamide | | Authors: | Ni, X, Kramer, J.S, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
9GZH
 
 | | PDGFRA-T674I in complex with covalent ponatinib derivative 19 | | Descriptor: | 3-(2-imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-~{N}-[4-(propanoylamino)-3-(trifluoromethyl)phenyl]benzamide, Platelet-derived growth factor receptor alpha | | Authors: | Teuber, A, Rauh, D, Mueller, M.P. | | Deposit date: | 2024-10-04 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, Synthesis, and SAR of Covalent KIT and PDGFRA Inhibitors─Exploring Their Potential in Targeting GIST.
J.Med.Chem., 68, 2025
|
|
7V8W
 
 | | Crystal structure of PsEst3 S128A variant complexed with malonate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MALONIC ACID, ... | | Authors: | Son, J, Kim, H, Kim, H.W. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
8DI4
 
 | | Discovery of MK-8189, a highly potent and selective PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 2-methyl-6-{[(1S,2S)-2-(5-methylpyridin-2-yl)cyclopropyl]methoxy}-N-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hayes, R.P, Yan, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia.
J.Med.Chem., 66, 2023
|
|
8FTG
 
 | | Biophysical and Structural Characterization of an Anti-Caffeine VHH Antibody | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-Caffeine VHH Antibody, CAFFEINE, ... | | Authors: | Horn, J.R, Smith, C.A, Sonneson, G.J, Walter, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Molecular recognition requires dimerization of a VHH antibody.
Mabs, 15, 2023
|
|
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
7NZI
 
 | | TrmB complex with SAH | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
9DYS
 
 | |
5X6X
 
 | |
6WHI
 
 | | Cryo-electron microscopy structure of the type I-F CRISPR RNA-guided surveillance complex bound to the anti-CRISPR AcrIF9 | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-04-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
