3MM5
 
 | | Dissimilatory sulfite reductase in complex with the substrate sulfite | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3UNK
 
 | | CDK2 in complex with inhibitor YL5-083 | | Descriptor: | 4-({4-[(2-chlorophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Cyclin-dependent kinase 2, PHOSPHATE ION | | Authors: | Zhu, J.-Y, Martin, M.P, Alam, R, Schonbrunn, E. | | Deposit date: | 2011-11-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3UNZ
 
 | | Aurora A in Complex with RPM1679 | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2-fluorophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3MAC
 
 | | crystal structure of GP41-derived protein complexed with fab 8062 | | Descriptor: | Fab8062, Transmembrane glycoprotein | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
3MPI
 
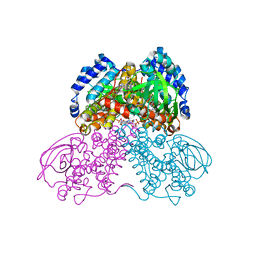 | | Structure of the glutaryl-coenzyme A dehydrogenase glutaryl-CoA complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, glutaryl-coenzyme A | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
3UOH
 
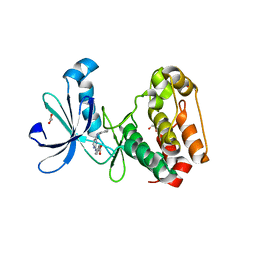 | | Aurora A in complex with RPM1722 | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2-bromophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8002 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3UOK
 
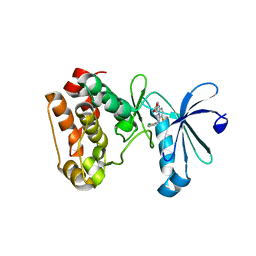 | | Aurora A in complex with YL5-81-1 | | Descriptor: | 4-({4-[(2-chlorophenyl)amino]-5-fluoropyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9506 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3ME1
 
 | |
3MI8
 
 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3M21
 
 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | Descriptor: | Probable tautomerase HP_0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3M6L
 
 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion
To be Published
|
|
3M85
 
 | |
3UOL
 
 | | Aurora A in complex with SO2-162 | | Descriptor: | 1,2-ETHANEDIOL, Aurora kinase A, N~4~-(2-chlorophenyl)-N~2~-[4-(1H-tetrazol-5-yl)phenyl]pyrimidine-2,4-diamine | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3V1Z
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP*G)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
3MBS
 
 | | Crystal structure of 8mer PNA | | Descriptor: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
3UX0
 
 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer Fusicoccin H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(hydroxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-gulopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Bartel, M, Anders, C, Higuchi, Y, Schumacher, B, Kato, N, Ottmann, C. | | Deposit date: | 2011-12-03 | | Release date: | 2013-01-02 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
3OL0
 
 | |
3P1R
 
 | | Crystal structure of human 14-3-3 sigma C38V/N166H in complex with TASK-3 peptide | | Descriptor: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CALCIUM ION, ... | | Authors: | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | Deposit date: | 2010-09-30 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
4AM6
 
 | | C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, SULFATE ION | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
402D
 
 | | 5'-R(*CP*GP*CP*CP*AP*GP*CP*G)-3' | | Descriptor: | RNA(5'-R(*CP*GP*CP*CP*AP*GP*CP*G)-3') | | Authors: | Jang, S.B, Hung, L.-W, Chi, Y.-I, Holbrook, E.L, Carter, R, Holbrook, S.R. | | Deposit date: | 1998-06-04 | | Release date: | 1998-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an RNA internal loop consisting of tandem C-A+ base pairs.
Biochemistry, 37, 1998
|
|
4A0Z
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | Descriptor: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
3OWG
 
 | | Crystal structure of vaccinia virus Polyadenylate polymerase(vp55) | | Descriptor: | Poly(A) polymerase catalytic subunit | | Authors: | Li, C, Li, H, Zhou, S, Gershon, P.D, Poulos, T.L. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Domain-level rocking motion within a polymerase that translocates on single-stranded nucleic acid.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3P08
 
 | |
3P0L
 
 | | Human steroidogenic acute regulatory protein | | Descriptor: | Steroidogenic acute regulatory protein, mitochondrial | | Authors: | Lehtio, L, Siponen, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
4AMS
 
 | |
