4WHY
 
 | |
4WQ7
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - "as isolated" form | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
3CZ6
 
 | | Crystal Structure of the Rap1 C-terminus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA-binding protein RAP1 | | Authors: | Feeser, E.A, Wolberger, C. | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants.
J.Mol.Biol., 380, 2008
|
|
1LKV
 
 | |
1KPA
 
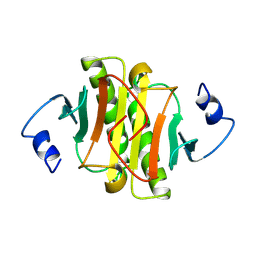 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPB
 
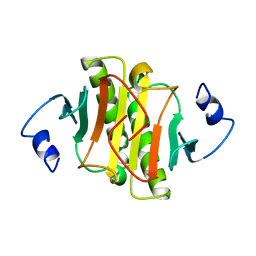 | | PKCI-1-APO | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
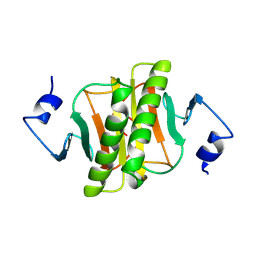 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KIB
 
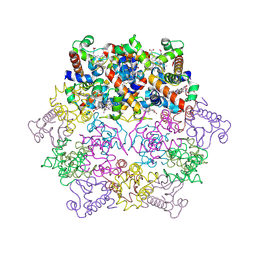 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KNK
 
 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
6THL
 
 | | Crystal structure of the complex between RTT106 and BCD1 | | Descriptor: | Box C/D snoRNA protein 1, Rtt106p | | Authors: | Charron, C, Bragantini, B, Manival, X, Charpentier, B. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The box C/D snoRNP assembly factor Bcd1 interacts with the histone chaperone Rtt106 and controls its transcription dependent activity.
Nat Commun, 12, 2021
|
|
1TRL
 
 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
2IIH
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
2IUM
 
 | |
1U2L
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P1) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2J
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P21 21 21) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2K
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (I41) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6TOP
 
 | | Structure of the PorE C-terminal domain, a protein of T9SS from Porphyromonas gingivalis | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, OmpA family protein, SODIUM ION | | Authors: | Trinh, T.N, Cambillau, C, Roussel, A, Leone, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Sci Rep, 10, 2020
|
|
8HIG
 
 | |
6TGK
 
 | | Domain swapped E6AP C-lobe dimer | | Descriptor: | ACETATE ION, CALCIUM ION, Ubiquitin-protein ligase E3A | | Authors: | Ries, L.K, Feiler, C, Lowe, L.D, Liess, A.K.L, Lorenz, S. | | Deposit date: | 2019-11-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the catalytic C-lobe of the HECT-type ubiquitin ligase E6AP.
Protein Sci., 29, 2020
|
|
2J4Z
 
 | | Structure of Aurora-2 in complex with PHA-680626 | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)-N-[5-(2-THIENYLACETYL)-1,5-DIHYDROPYRROLO[3,4-C]PYRAZOL-3-YL]BENZAMIDE, ARSENIC, SERINE THREONINE-PROTEIN KINASE 6 | | Authors: | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | Deposit date: | 2006-09-08 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile.
J. Med. Chem., 49, 2006
|
|
6TJT
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFC peptide | | Descriptor: | 1,2-ETHANEDIOL, Neuropilin-2, SULFATE ION, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFC peptide
To Be Published
|
|
4W92
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, C-di-AMP ribsoswitch, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
6TLJ
 
 | |
1UD0
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL 10-kDA SUBDOMAIN OF HSC70 | | Descriptor: | 70 kDa heat-shock-like protein, SODIUM ION | | Authors: | Chou, C.C, Forouhar, F, Yeh, Y.H, Wang, C, Hsiao, C.D. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the C-terminal 10-kDa subdomain of Hsc70
J.BIOL.CHEM., 278, 2003
|
|
8AJZ
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
