4HVP
 
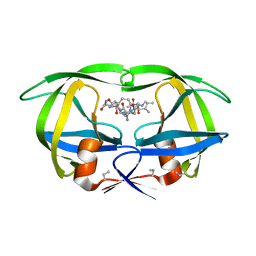 | | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Angstroms resolution | | Descriptor: | HIV-1 PROTEASE, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide | | Authors: | Miller, M, Schneider, J, Sathyanarayana, B.K, Toth, M.V, Marshall, G.R, Clawson, L, Selk, L, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1989-08-08 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution.
Science, 246, 1989
|
|
3CPP
 
 | |
5EBX
 
 | |
5ANA
 
 | |
2I1B
 
 | |
4I1B
 
 | | FUNCTIONAL IMPLICATIONS OF INTERLEUKIN-1BETA BASED ON THE THREE-DIMENSIONAL STRUCTURE | | Descriptor: | INTERLEUKIN-1 BETA | | Authors: | Veerapandian, B, Poulos, T.L, Gilliland, G.L, Raag, R, Svensson, L.A, Masui, Y, Hirai, Y. | | Deposit date: | 1990-03-27 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional implications of interleukin-1 beta based on the three-dimensional structure.
Proteins, 12, 1992
|
|
4PEP
 
 | |
1P05
 
 | |
4INS
 
 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
2AIT
 
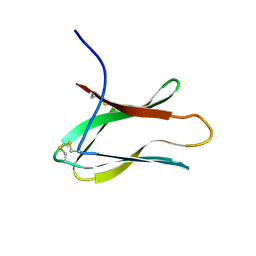 | | DETERMINATION OF THE COMPLETE THREE-DIMENSIONAL STRUCTURE OF THE ALPHA-AMYLASE INHIBITOR TENDAMISTAT IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE AND DISTANCE GEOMETRY | | Descriptor: | TENDAMISTAT | | Authors: | Kline, A.D, Braun, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1989-05-24 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the complete three-dimensional structure of the alpha-amylase inhibitor tendamistat in aqueous solution by nuclear magnetic resonance and distance geometry.
J.Mol.Biol., 204, 1988
|
|
2HLA
 
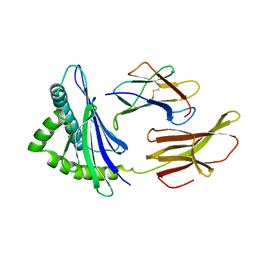 | |
3PEP
 
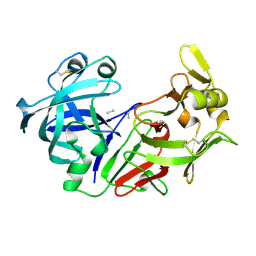 | |
1LYD
 
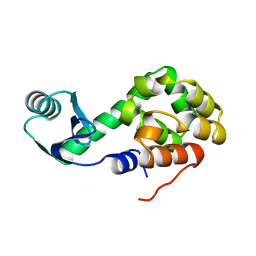 | |
4FAB
 
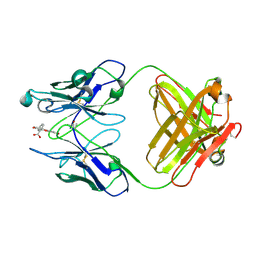 | | THREE-DIMENSIONAL STRUCTURE OF A FLUORESCEIN-FAB COMPLEX CRYSTALLIZED IN 2-METHYL-2,4-PENTANEDIOL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, IGG2A-KAPPA 4-4-20 FAB (HEAVY CHAIN), ... | | Authors: | Herron, J.N, He, X, Mason, M.L, Vossjunior, E.W, Edmundson, A.B. | | Deposit date: | 1989-04-10 | | Release date: | 1990-07-15 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a fluorescein-Fab complex crystallized in 2-methyl-2,4-pentanediol.
Proteins, 5, 1989
|
|
5DFR
 
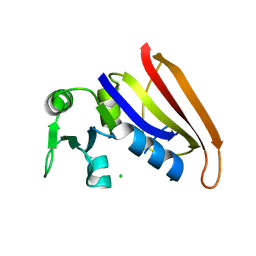 | |
6DFR
 
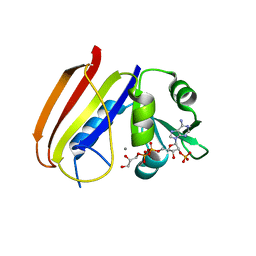 | |
6ACN
 
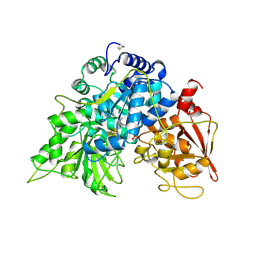 | |
2DHF
 
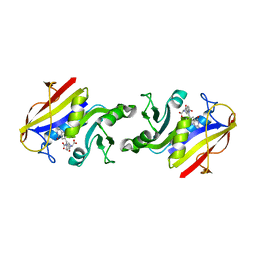 | |
5ACN
 
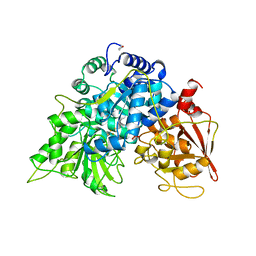 | |
5PEP
 
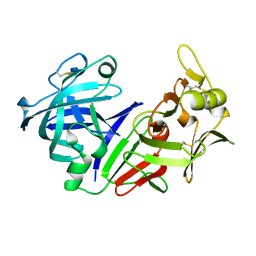 | | X-RAY ANALYSES OF ASPARTIC PROTEASES. II. THREE-DIMENSIONAL STRUCTURE OF THE HEXAGONAL CRYSTAL FORM OF PORCINE PEPSIN AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | PEPSIN | | Authors: | Cooper, J.B, Khan, G, Taylor, G, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-05-30 | | Release date: | 1990-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution.
J.Mol.Biol., 214, 1990
|
|
2CLA
 
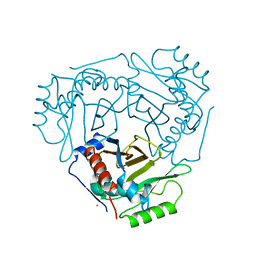 | |
2CHY
 
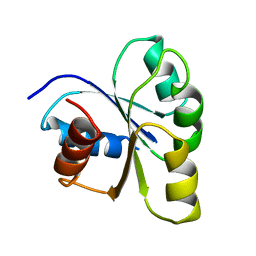 | | THREE-DIMENSIONAL STRUCTURE OF CHEY, THE RESPONSE REGULATOR OF BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Mottonen, J.M, Stock, A.M, Stock, J.B, Schutt, C.E. | | Deposit date: | 1990-05-17 | | Release date: | 1990-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis.
Nature, 337, 1989
|
|
1CLA
 
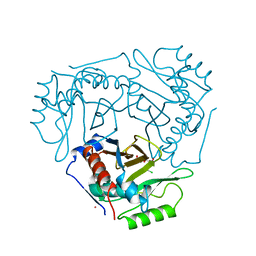 | |
1DHF
 
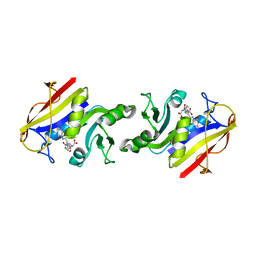 | |
1R1A
 
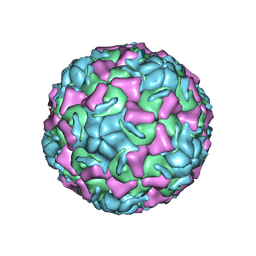 | | CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS SEROTYPE 1A (HRV1A) | | Descriptor: | HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Kim, S, Rossmann, M.G. | | Deposit date: | 1989-03-15 | | Release date: | 1990-07-15 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human rhinovirus serotype 1A (HRV1A).
J.Mol.Biol., 210, 1989
|
|
