3E6P
 
 | | Crystal structure of human meizothrombin desF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANYL-N-[(1S)-4-{[(Z)-AMINO(IMINO)METHYL]AMINO}-1-(CHLOROACETYL)BUTYL]-L-PROLINAMIDE, Prothrombin, ... | | Authors: | Papaconstantinou, M.E, Gandhi, P, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2008-08-15 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Na(+) binding to meizothrombin desF1.
Cell.Mol.Life Sci., 65, 2008
|
|
2OVP
 
 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
1EFC
 
 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
1MNX
 
 | |
4FXH
 
 | |
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.14885437 Å) | | Cite: | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
2OVR
 
 | | Structure of the Skp1-Fbw7-CyclinEdegN complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
1YVT
 
 | | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35) | | Descriptor: | CARBON MONOXIDE, GLYCEROL, Hemoglobin alpha chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35)
To be Published
|
|
2OS3
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, COBALT (II) ION, Peptide deformylase | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Parh, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
1ZKC
 
 | | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b | | Descriptor: | BETA-MERCAPTOETHANOL, Peptidyl-prolyl cis-trans isomerase like 2 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2OS0
 
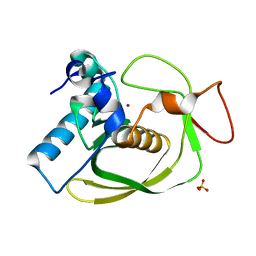 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
1YVQ
 
 | | The low salt (PEG) crystal structure of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5) | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5)
To be Published
|
|
1ZNA
 
 | |
7YRD
 
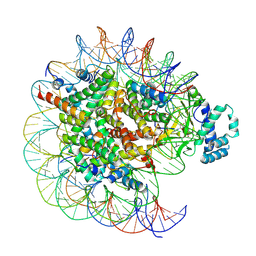 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7YRG
 
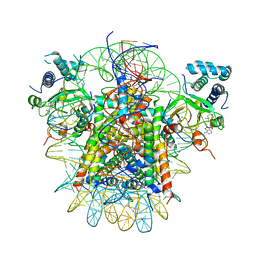 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
4TYV
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ3
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ1
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ5
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TTG
 
 | | Beta-galactosidase (E. coli) in the presence of potassium chloride. | | Descriptor: | Beta-galactosidase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2014-06-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating factors important for monovalent cation selectivity in enzymes: E. coli beta-galactosidase as a model.
Phys Chem Chem Phys, 17, 2015
|
|
4RUL
 
 | | Crystal structure of full-length E.Coli topoisomerase I in complex with ssDNA | | Descriptor: | DNA topoisomerase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Chen, B, Tse-Dinh, Y.C. | | Deposit date: | 2014-11-20 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I.
Nucleic Acids Res., 43, 2015
|
|
4V69
 
 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4UDT
 
 | | T cell receptor (TRAV22,TRBV7-9) structure | | Descriptor: | GLYCEROL, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
4RWB
 
 | | Racemic influenza M2-TM crystallized from monoolein lipidic cubic phase | | Descriptor: | Matrix protein 2, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
