5F0C
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium tuberculosis in complex with compound 4 at 1.87A resolution | | Descriptor: | 3-cyclopentyl-~{N}-(3-piperidin-1-ylphenyl)propanamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
3EJT
 
 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog:(5R)-5-[2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (1S,2R,5R,8R,8aR)-5-[2-(4-tert-butylphenyl)ethyl]octahydroindolizine-1,2,8-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
5EZ2
 
 | | Sandercyanin Fluorescent Protein (SFP) | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Ramaswamy, S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9G0D
 
 | | Structure of human Mical1 bMERB_V978A domain:Rab10 complex. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-10, ... | | Authors: | Rai, A, Goody, R.S. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural insights into the auto-inhibited state of Mical1 and its activation by Rab8
Elife, 2024
|
|
3EKN
 
 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[4-(1-methylethyl)piperazin-1-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2ORV
 
 | | human Thymidine Kinase 1 in complex with TP4A | | Descriptor: | P1-(5'-ADENOSYL)P4-(5'-(2'-DEOXY-THYMIDYL))TETRAPHOSPHATE, Thymidine kinase, ZINC ION | | Authors: | Segura-Pena, D, Lutz, S, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2007-02-04 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of ATP to TK1-like Enzymes Is Associated with a Conformational Change in the Quaternary Structure.
J.Mol.Biol., 369, 2007
|
|
3MAM
 
 | | A molecular switch changes the low to the high affinity state in the substrate binding protein AfProX | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Osmoprotection protein (ProX), ... | | Authors: | Tschapek, B, Pittelkow, M, Bremer, E, Schmitt, L, Smits, S.H. | | Deposit date: | 2010-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arg149 Is Involved in Switching the Low Affinity, Open State of the Binding Protein AfProX into Its High Affinity, Closed State.
J.Mol.Biol., 411, 2011
|
|
4GCJ
 
 | | CDK2 in complex with inhibitor RC-3-89 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
8IOZ
 
 | | Crystal structure of transaminase | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-01-24 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Protein engineering of transaminase facilitating enzyme cascade reaction for the biosynthesis of azasugars.
Iscience, 27, 2024
|
|
3EO4
 
 | | The crystal structure of a domain from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Tan, K, Hatzos, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a domain from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
5F25
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 4. | | Descriptor: | 4-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)benzamide, BRD9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
9G6T
 
 | | p53-Y220C Core Domain Covalently Bound to 5-Chloro-6-methylpyrazine-2-carbonitrile Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranyl-6-methyl-pyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
3UVD
 
 | | Crystal Structure of the bromodomain of human Transcription activator BRG1 (SMARCA4) in complex with N-Methyl-2-pyrrolidone | | Descriptor: | 1-methylpyrrolidin-2-one, Transcription activator BRG1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
6SRH
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2117 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-09-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2117
To Be Published
|
|
3MPD
 
 | |
8IAJ
 
 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, Protein ORM2, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
6DHL
 
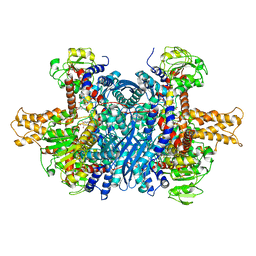 | | Bovine glutamate dehydrogenase complexed with epicatechin-3-gallate (ECG) | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site.
J. Biol. Chem., 286, 2011
|
|
6ZOU
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB333 | | Descriptor: | 11-methyl-~{N}-[(2~{S},3~{R})-1-[[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]dodecanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
3UVK
 
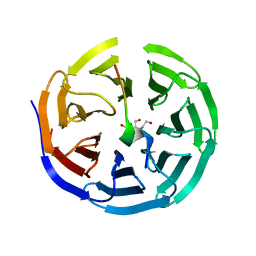 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase MLL2, SULFATE ION, ... | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
5F6Y
 
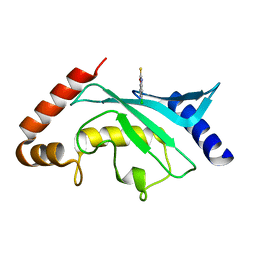 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8IAM
 
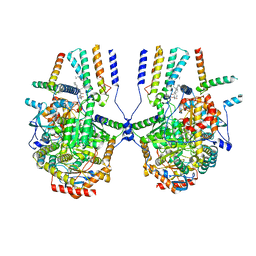 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3D) complex | | Descriptor: | Chimera of Long chain base biosynthesis protein 1 and Serine palmitoyltransferase 1, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
9FXZ
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with 4-(bromophenyl)phthalazinone D-galactal ligand | | Descriptor: | 4-(4-bromophenyl)-2-[[(2~{R},3~{R},4~{R})-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2~{H}-pyran-4-yl]oxymethyl]phthalazin-1-one, CHLORIDE ION, Galectin-8 | | Authors: | Van Klaveren, S, Hakansson, M, Diehl, C, Nilsson, N.J. | | Deposit date: | 2024-07-02 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Galectin-8N-Selective 4-Halophenylphthalazinone-Galactals Double pi-Stack in a Unique Pocket.
Acs Med.Chem.Lett., 15, 2024
|
|
1MBD
 
 | |
1EOM
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 WITH A BIANTENNARY COMPLEX OCTASACCHARIDE | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
6ZP6
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB334 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
