4O3C
 
 | | Crystal structure of the GLUA2 ligand-binding domain in complex with L-aspartate at 1.50 A resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, K, Kaern, A.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
2BVT
 
 | | The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. | | Descriptor: | BETA-1,4-MANNANASE, CACODYLATE ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Le Nours, J, Anderson, L, Stoll, D, Stalbrand, H, Lo Leggio, L. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Characterization of a Modular Endo-Beta-1,4-Mannanase from Cellulomonas Fimi
Biochemistry, 44, 2005
|
|
4O6K
 
 | | The crystal structure of zebrafish IL-22 | | Descriptor: | Interleukin 22 | | Authors: | Siupka, P, Hamming, O.J, Fretaud, M, Luftalla, G, Levraud, J.P, Hartmann, R. | | Deposit date: | 2013-12-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of zebrafish IL-22 reveals an evolutionary, conserved structure highly similar to that of human IL-22.
Genes Immun., 15, 2014
|
|
4IGT
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution | | Descriptor: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2012-12-18 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
2ANJ
 
 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
1AWH
 
 | | NOVEL COVALENT THROMBIN INHIBITOR FROM PLANT EXTRACT | | Descriptor: | 3-ACETOXY-17-(1-FORMYL-5-METHYL-3-OXO-HEX-4-ENYL)-16-HYDROXY-4,10,13,14-TETRAMETHYL-2,3,4,5,6,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYCLOPENTA[A]PHENANTHRENE-4-CARBOXYLIC ACID, ALPHA THROMBIN | | Authors: | Jhoti, H, Cleasby, A, Wonacott, A. | | Deposit date: | 1997-10-02 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel natural product 5,5-trans-lactone inhibitors of human alpha-thrombin: mechanism of action and structural studies.
Biochemistry, 37, 1998
|
|
1NXI
 
 | | Solution structure of Vibrio cholerae protein VC0424 | | Descriptor: | conserved hypothetical protein VC0424 | | Authors: | Ramelot, T.A, Ni, S, Goldsmith-Fischman, S, Cort, J.R, Honig, B, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-10 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Vibrio cholerae protein VC0424: a variation of the ferredoxin-like fold.
Protein Sci., 12, 2003
|
|
2BYY
 
 | | E.coli KAS I H298E Mutation | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE I, AMMONIUM ION | | Authors: | Olsen, J.G, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2005-08-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fatty acid synthesis. Role of active site histidines and lysine in Cys-His-His-type beta-ketoacyl-acyl carrier protein synthases.
FEBS J., 273, 2006
|
|
2BNY
 
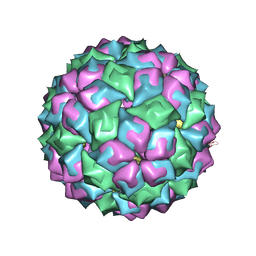 | | MS2 (N87A mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BGS
 
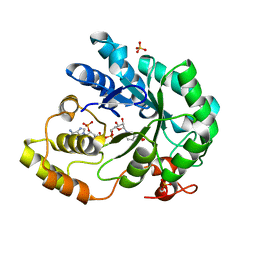 | | HOLO ALDOSE REDUCTASE FROM BARLEY | | Descriptor: | ALDOSE REDUCTASE, BICARBONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-05 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
4LP4
 
 | |
2BYZ
 
 | |
1BWQ
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
4M4L
 
 | | The structure of Cu T6 bovine insulin | | Descriptor: | COPPER (II) ION, Insulin | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2BYX
 
 | |
4IY6
 
 | | Crystal structure of the GLUA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and ME-CX516 at 1.72 A resolution | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-01-28 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J0N
 
 | | Crystal structure of a manganese dependent isatin hydrolase | | Descriptor: | CALCIUM ION, Isatin hydrolase B, MANGANESE (II) ION, ... | | Authors: | Bjerregaard-Andersen, K, Sommer, T, Jensen, J.K, Jochimsen, B, Etzerodt, M, Morth, J.P. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A proton wire and water channel revealed in the crystal structure of isatin hydrolase.
J.Biol.Chem., 289, 2014
|
|
2BZ3
 
 | | Structure of E.coli KAS I H298E mutant in complex with C12 fatty acid | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE I, AMMONIUM ION, LAURIC ACID, ... | | Authors: | Olsen, J.G, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2005-08-10 | | Release date: | 2006-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fatty acid synthesis. Role of active site histidines and lysine in Cys-His-His-type beta-ketoacyl-acyl carrier protein synthases.
FEBS J., 273, 2006
|
|
2BQ5
 
 | | MS2 (N87AE89K mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-27 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BQV
 
 | | HIV-1 protease in complex with inhibitor AHA455 | | Descriptor: | 6-AMINO HEXANOIC ACID, HIV-1 PROTEASE | | Authors: | Unge, T, Ekegren, J.K, Schenk, H.V, Zreik Safa, M, Wallberg, H, Samuelsson, B, Hallberg, A. | | Deposit date: | 2005-04-28 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A New Class of HIV-1 Protease Inhibitors Containing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 48, 2005
|
|
4IY5
 
 | | Crystal structure of the glua2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and CX516 at 2.0 A resolution | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-01-28 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1LV3
 
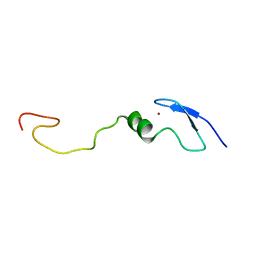 | | Solution NMR Structure of Zinc Finger Protein yacG from Escherichia coli. Northeast Structural Genomics Consortium Target ET92. | | Descriptor: | HYPOTHETICAL PROTEIN YacG, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-24 | | Release date: | 2002-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins.
Proteins, 49, 2002
|
|
1BWP
 
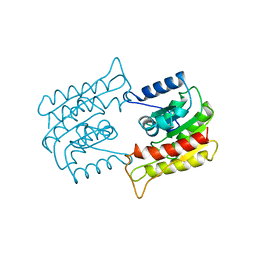 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
4N07
 
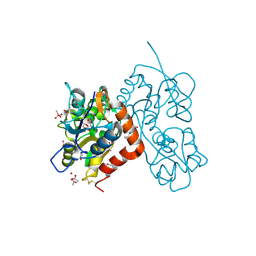 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
1BWR
 
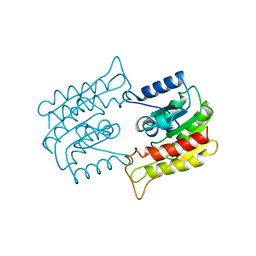 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
