1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HLE
 
 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
3GE2
 
 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
3GHD
 
 | | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Descriptor: | a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Authors: | Dong, A, Xu, X, Chruszcz, M, Brown, G, Proudfoot, M, Edwards, A.M, Joachimiak, A, Minor, W, Savchenko, A, Yaleunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain
To be Published
|
|
3GRP
 
 | |
6TCX
 
 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
3EFG
 
 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
3BOR
 
 | | Crystal structure of the DEADc domain of human translation initiation factor 4A-2 | | Descriptor: | Human initiation factor 4A-II | | Authors: | Dimov, S, Hong, B, Tempel, W, MacKenzie, F, Karlberg, T, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3IDO
 
 | |
3IEZ
 
 | | Crystal structure of the RasGAP C-terminal (RGC) domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-23 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the RasGAP C-terminal (RGC) domain
of IQGAP2
To be Published
|
|
4IDI
 
 | | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420 | | Descriptor: | GLYCEROL, Oryza sativa Rurm1-related | | Authors: | Wernimont, A.K, Tempel, W, Lew, J, Walker, J, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Bountra, C, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-12 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420
To be Published
|
|
4LQC
 
 | | The crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry. | | Descriptor: | TcpB | | Authors: | Snyder, G.A, Smith, P, Fresquez, T, Cirl, C, Jiang, J, Snyder, N, Luchetti, T, Miethke, T, Xiao, T.S. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry.
J.Biol.Chem., 289, 2014
|
|
3HE1
 
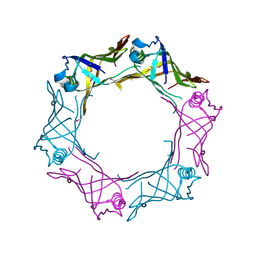 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
3HIL
 
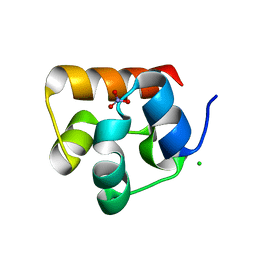 | | SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1) | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 1, NITRATE ION | | Authors: | Walker, J.R, Yermekbayeva, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1).
To be Published
|
|
3KKA
 
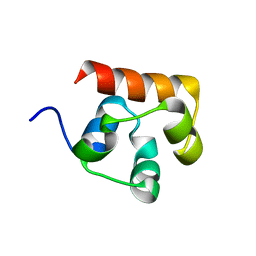 | | Co-crystal structure of the sam domains of EPHA1 AND EPHA2 | | Descriptor: | CHLORIDE ION, EPHRIN TYPE-A RECEPTOR 1, EPHRIN TYPE-A RECEPTOR 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Co-Crystal Structure of the SAM Domains of Human Ephrin Type-A Receptor 1 and Human Ephrin Type-A Receptor 2
To be Published
|
|
3KZV
 
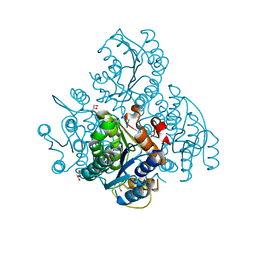 | | The crystal structure of a cytoplasmic protein with unknown function from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Uncharacterized oxidoreductase YIR035C | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a cytoplasmic protein with unknown function from Saccharomyces cerevisiae
To be Published
|
|
3KUZ
 
 | | Crystal structure of the ubiquitin like domain of PLXNC1 | | Descriptor: | Plexin-C1, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Li, B, Tempel, W, Tong, Y, Guan, X, Zhong, N, Crombet, L, MacKenzie, F, Buck, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ubiquitin like domain of PLXNC1
to be published
|
|
3LT3
 
 | | Crystal structure of Rv3671c from M. tuberculosis H37Rv, Ser343Ala mutant, inactive form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2010-02-14 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
3LZK
 
 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
3J9C
 
 | | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Protective antigen PA-63 | | Authors: | Jiang, J, Pentelute, B.L, Collier, R.J, Zhou, Z.H. | | Deposit date: | 2014-12-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of anthrax protective antigen pore elucidates toxin translocation.
Nature, 521, 2015
|
|
1Q56
 
 | | NMR structure of the B0 isoform of the agrin G3 domain in its Ca2+ bound state | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-08-06 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding
Structure, 12, 2004
|
|
1PZ8
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1PZ9
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
3KWV
 
 | |
1PZ7
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
