3BTV
 
 | |
3BTU
 
 | |
3BTS
 
 | |
7RGT
 
 | | The crystal structure of RocC, containing FinO domain, 1-126 | | Descriptor: | Repressor of competence, RNA Chaperone, SULFATE ION | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGS
 
 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
1R1U
 
 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the apo-form | | Descriptor: | repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
8B4A
 
 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7SA1
 
 | | LRR-F-Box plant ubiquitin ligase | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, F-box/LRR-repeat MAX2 homolog, ... | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2021-09-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling.
Nat.Plants, 8, 2022
|
|
6UH8
 
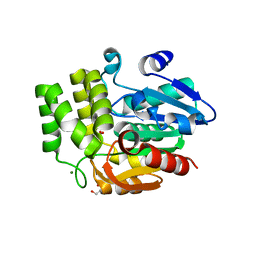 | | Crystal structure of DAD2 N242I mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Decreased Apical Dominance 2, GLYCEROL, ... | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
6UH9
 
 | | Crystal structure of DAD2 D166A mutant | | Descriptor: | Decreased Apical Dominance 2, TETRAETHYLENE GLYCOL | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
3VXK
 
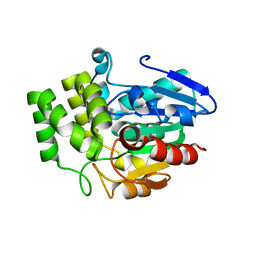 | | Crystal structure of OsD14 | | Descriptor: | Dwarf 88 esterase | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
3BTL
 
 | |
1K9H
 
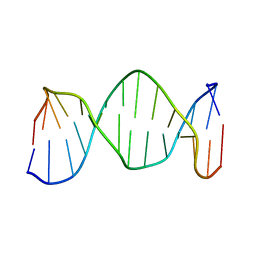 | | NMR structure of DNA TGTGAGCGCTCACA | | Descriptor: | 5'-D(*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*A)-3' | | Authors: | Kaluarachchi, K, Gorenstein, D.G, Luxon, B.A. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How Do Proteins Recognize DNA? Solution Structure and Local Conformational Dynamics of Lac Operators by 2D NMR
J.Biomol.Struct.Dyn., Conversation 11, 2000
|
|
4V4P
 
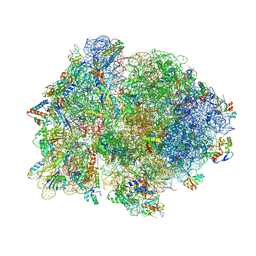 | | Crystal structure of 70S ribosome with thrS operator and tRNAs. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Romby, P, Rees, B, Schulze-Briese, C, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D, Yusupova, G, Yusupov, M. | | Deposit date: | 2005-01-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
Science, 308, 2005
|
|
6EJZ
 
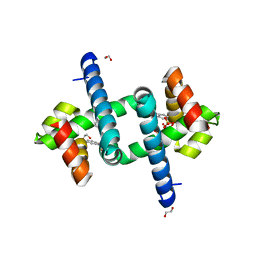 | | Tryptophan Repressor TrpR from E.coli variant S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, SULFATE ION, ... | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
2DI3
 
 | |
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | Descriptor: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | Authors: | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2013-08-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
8JFV
 
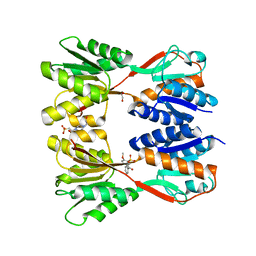 | | Crystal structure of Catabolite repressor acivator from E. coli in complex with sulisobenzone | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-oxidanyl-5-(phenylcarbonyl)benzenesulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Neetu, N, Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Sulisobenzone is a potent inhibitor of the global transcription factor Cra.
J.Struct.Biol., 215, 2023
|
|
6L5K
 
 | | ARF5 Aux/IAA17 Complex | | Descriptor: | Auxin response factor 5, Auxin-responsive protein IAA17 | | Authors: | Ryu, K.S, Suh, J.Y, Cha, S.Y, Kim, Y.I, Park, C.K. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Determinants of PB1 Domain Interactions in Auxin Response Factor ARF5 and Repressor IAA17.
J.Mol.Biol., 432, 2020
|
|
4WKM
 
 | | AmpR effector binding domain from Citrobacter freundii bound to UDP-MurNAc-pentapeptide | | Descriptor: | ALA-FGA-API-DAL-DAL, GLYCEROL, LysR family transcriptional regulator, ... | | Authors: | Vadlamani, G, Reeve, T.M, Mark, B.L. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The beta-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide.
J.Biol.Chem., 290, 2015
|
|
5X9P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
7C7E
 
 | | Crystal structure of C terminal domain of Escherichia coli DgoR | | Descriptor: | Putative DNA-binding transcriptional regulator, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Lin, W. | | Deposit date: | 2020-05-25 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural and Functional Analyses of the Transcription Repressor DgoR From Escherichia coli Reveal a Divalent Metal-Containing D-Galactonate Binding Pocket.
Front Microbiol, 11, 2020
|
|
1PPF
 
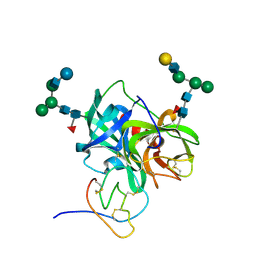 | | X-RAY CRYSTAL STRUCTURE OF THE COMPLEX OF HUMAN LEUKOCYTE ELASTASE (PMN ELASTASE) AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR | | Descriptor: | HUMAN LEUKOCYTE ELASTASE, TURKEY OVOMUCOID INHIBITOR (OMTKY3), alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bode, W, Wei, A-Z. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the complex of human leukocyte elastase (PMN elastase) and the third domain of the turkey ovomucoid inhibitor.
EMBO J., 5, 1986
|
|
3N00
 
 | |
7Z36
 
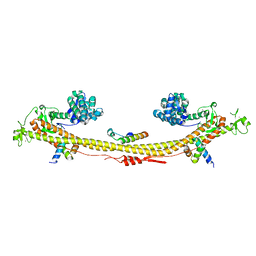 | | Crystal structure of the KAP1 tripartite motif in complex with the ZNF93 KRAB domain | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta,Isoform 2 of Transcription intermediary factor 1-beta, SMARCAD1 CUE1 domain, ZINC ION, ... | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional mapping of the KRAB-KAP1 repressor complex.
Embo J., 41, 2022
|
|
