5W4C
 
 | | Crystal structure of thioredoxin reductase from Cryptococcus neoformans in complex with FAD (FO conformation) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo-Chaucanes, C.P, Valadares, N.F, Felipe, M.S.S, Barbosa, J.A.R.G. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of thioredoxin reductase from Cryptococcus neoformans in complex with FAD (FO conformation)
To be published
|
|
6Q3X
 
 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclohexane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
2JYF
 
 | |
3SCF
 
 | | Fe(II)-HppE with S-HPP and NO | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, Epoxidase, FE (II) ION, ... | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
2IID
 
 | | Structure of L-amino acid oxidase from Calloselasma rhodostoma in complex with L-phenylalanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino-acid oxidase, ... | | Authors: | Moustafa, I.M, Foster, S, Lyubimov, A.Y, Vrielink, A. | | Deposit date: | 2006-09-27 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of LAAO from Calloselasma rhodostoma with an L-Phenylalanine Substrate: Insights into Structure and Mechanism
J.Mol.Biol., 364, 2006
|
|
3SCG
 
 | | Fe(II)-HppE with R-HPP | | Descriptor: | Epoxidase, FE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
2HN8
 
 | | Structural characterization and oligomerization of PB1-F2, a pro-apoptotic influenza A virus protein | | Descriptor: | Protein PB1-F2 | | Authors: | Bruns, K, Studtrucker, N, Sharma, A, Fossen, T, Mitzner, D, Eissmann, A, Tessmer, U, Roder, R, Henklein, P, Wray, V, Schubert, U. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterization and oligomerization of PB1-F2, a pro-apoptotic influenza A virus protein.
J.Biol.Chem., 282, 2007
|
|
4FXE
 
 | |
2KI7
 
 | | The solution structure of RPP29-RPP21 complex from Pyrococcus furiosus | | Descriptor: | Ribonuclease P protein component 1, Ribonuclease P protein component 4, ZINC ION | | Authors: | Xu, Y, Foster, M.P. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions.
J.Mol.Biol., 393, 2009
|
|
4FXH
 
 | |
4FXI
 
 | |
2QPF
 
 | | Crystal Structure of Mouse Transthyretin | | Descriptor: | Transthyretin | | Authors: | Reixach, N, Foss, T.R, Santelli, E, Pascual, J, Kelly, J.W, Buxbaum, J.N. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human-Murine Transthyretin Heterotetramers Are Kinetically Stable and Non-amyloidogenic: A LESSON IN THE GENERATION OF TRANSGENIC MODELS OF DISEASES INVOLVING OLIGOMERIC PROTEINS.
J.Biol.Chem., 283, 2008
|
|
1KEZ
 
 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7NE2
 
 | | Crystal structure of class I SFP aldolase YihT from Salmonella enterica with SFP/ DHAP (Schiff base complex with active site Lys193) | | Descriptor: | (2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)-6-phosphonooxy-hexane-1-sulfonic acid, Sulfofructosephosphate aldolase, [(~{E})-2,3-bis(oxidanyl)prop-1-enyl] dihydrogen phosphate | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
4AWL
 
 | | The NF-Y transcription factor is structurally and functionally a sequence specific histone | | Descriptor: | HSP70 PROMOTER FRAGMENT, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT ALPHA, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT BETA, ... | | Authors: | Nardini, M, Gnesutta, N, Donati, G, Gatta, R, Forni, C, Fossati, A, Vonrhein, C, Moras, D, Romier, C, Mantovani, R, Bolognesi, M. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Sequence-Specific Transcription Factor NF-Y Displays Histone-Like DNA Binding and H2B-Like Ubiquitination.
Cell(Cambridge,Mass.), 152, 2013
|
|
4CBW
 
 | | Crystal structure of Plasmodium berghei actin I with D-loop from muscle actin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
2WBN
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 | | Descriptor: | TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
2WC9
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 with bound Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
1PQS
 
 | | Solution structure of the C-terminal OPCA domain of yCdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Leitner, D, Wahl, M, Labudde, D, Diehl, A, Schmieder, P, Pires, J.R, Fossi, M, Leidert, M, Krause, G, Oschkinat, H. | | Deposit date: | 2003-06-19 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an N-terminally truncated version of the yeast CDC24p PB1 domain shows a different beta-sheet topology.
Febs Lett., 579, 2005
|
|
4CBU
 
 | | Crystal structure of Plasmodium falciparum actin I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
2XCS
 
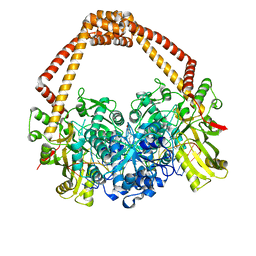 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCT
 
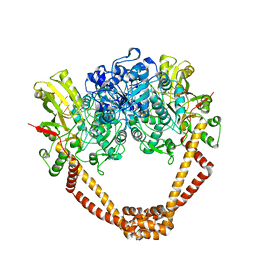 | | The twinned 3.35A structure of S. aureus Gyrase complex with Ciprofloxacin and DNA | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 5'-D(AP*GP*CP*CP*GP*TP*AP*G)-3', 5'-D(GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3', ... | | Authors: | Bax, B.D, Chan, P, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
4CBX
 
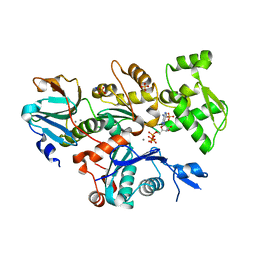 | | Crystal structure of Plasmodium berghei actin II | | Descriptor: | ACTIN-2, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
1OQK
 
 | | Structure of Mth11: A homologue of human RNase P protein Rpp29 | | Descriptor: | conserved protein MTH11 | | Authors: | Boomershine, W.P, McElroy, C.A, Tsai, H, Gopalan, V, Foster, M.P. | | Deposit date: | 2003-03-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
7PQH
 
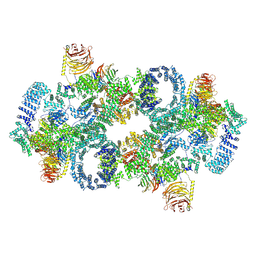 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
