4M5Q
 
 | | High-resolution apo influenza 2009 H1N1 endonuclease structure | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Polymerase PA | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
3TG5
 
 | | Structure of SMYD2 in complex with p53 and SAH | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
4E6C
 
 | | p38a-perifosine Complex | | Descriptor: | (1,1-dimethylpiperidin-1-ium-4-yl) octadecyl hydrogen phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
4E8A
 
 | | The crystal structure of p38a MAP kinase in complex with PIA24 | | Descriptor: | (1R,2S,3R,4S,6S)-6-(cyclohexylmethoxy)-2,3,4-trihydroxycyclohexyl (2R)-2-methoxy-3-(octadecyloxy)propyl hydrogen (S)-phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-20 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
4M4Q
 
 | | 6-(4-fluorophenyl)-3-hydroxy-5-[4-(1H-1,2,3,4-tetrazol-5-yl)phenyl] -1,2-dihydropyridin-2-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 6-(4-fluorophenyl)-5-[4-(1H-tetrazol-5-yl)phenyl]pyridine-2,3-diol, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
6QTJ
 
 | | Crystal structure of human CDK8/CYCC in complex with BI 919811 | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, ~{N},~{N}-dimethyl-2-[4-[4-(2,6-naphthyridin-4-yl)phenyl]pyrazol-1-yl]ethanamide | | Authors: | Boettcher, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
3L8S
 
 | | Human p38 MAP Kinase in Complex with CP-547632 | | Descriptor: | 3-[(4-bromo-2,6-difluorobenzyl)oxy]-5-{[(4-pyrrolidin-1-ylbutyl)carbamoyl]amino}isothiazole-4-carboxamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2010-01-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fluorophore labeling of the glycine-rich loop as a method of identifying inhibitors that bind to active and inactive kinase conformations.
J.Am.Chem.Soc., 132, 2010
|
|
2FSM
 
 | |
2FST
 
 | |
6R3S
 
 | | CRYSTAL STRUCTURE OF CDK8-CycC IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-[5-chloranyl-4-[(1~{S})-1-oxidanylethyl]pyridin-3-yl]-3,4-dihydro-2~{H}-1,8-naphthyridine-1-carboxamide, Cyclin-C, ... | | Authors: | Boettcher, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
7PMK
 
 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation I) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA helicase, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
2FSO
 
 | |
6N7S
 
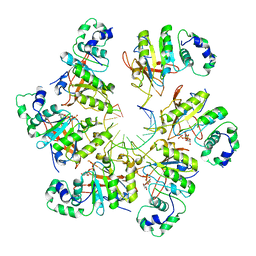 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form II) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6SIS
 
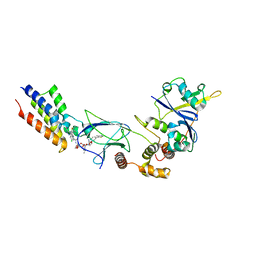 | | Crystal structure of macrocyclic PROTAC 1 in complex with the second bromodomain of human Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Hughes, S.J, Testa, A, Ciulli, A. | | Deposit date: | 2019-08-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Based Design of a Macrocyclic PROTAC.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2FSL
 
 | |
6QTG
 
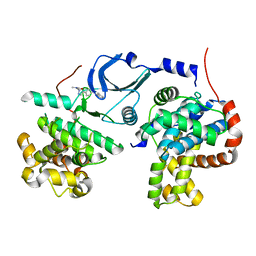 | | Crystal structure of human CDK8/CYCC in complex with BI-1347 | | Descriptor: | 2-[4-(4-isoquinolin-4-ylphenyl)pyrazol-1-yl]-~{N},~{N}-dimethyl-ethanamide, Cyclin-C, Cyclin-dependent kinase 8 | | Authors: | Boettcher, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
3FQI
 
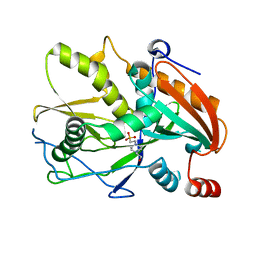 | | Crystal Structure of the Mouse Dom3Z | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Protein Dom3Z | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458, 2009
|
|
3FQJ
 
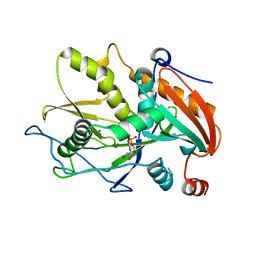 | | Crystal Structure of the Mouse Dom3Z in Complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein Dom3Z | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458, 2009
|
|
7TYR
 
 | |
2BAK
 
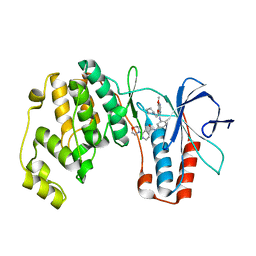 | | p38alpha MAP kinase bound to MPAQ | | Descriptor: | Mitogen-activated protein kinase 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Breed, J, Pauptit, R.A, Read, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAJ
 
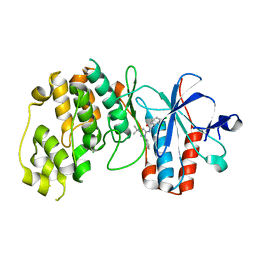 | | p38alpha bound to pyrazolourea | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2,3-dichlorophenyl)urea, Mitogen-activated protein kinase 14 | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Breed, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAQ
 
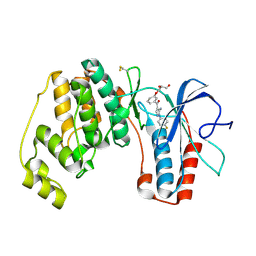 | | p38alpha bound to Ro3201195 | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Breed, J, Read, J, Tucker, J, Norman, R.A. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
6NEL
 
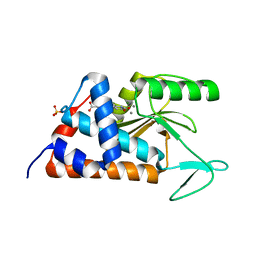 | | 4-(2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzoic acid bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 4-[2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl]benzoic acid, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl and Arylalkyl Substituted 3-Hydroxypyridin-2(1H)-ones: Synthesis and Evaluation as Inhibitors of Influenza A Endonuclease.
Chemmedchem, 14, 2019
|
|
5M2N
 
 | |
8OSL
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
