3PJG
 
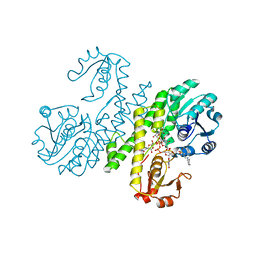 | | Crystal structure of UDP-glucose dehydrogenase from Klebsiella pneumoniae complexed with product UDP-glucuronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
5TWA
 
 | | Crystal structure of Geodia cydonium BHP2 in complex with Lubomirskia baicalensis Bak-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAK-2 protein, ... | | Authors: | Caria, S, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2016-11-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into an evolutionarily ancient programmed cell death regulator - the crystal structure of marine sponge BHP2 bound to LB-Bak-2.
Cell Death Dis, 8, 2017
|
|
5TTO
 
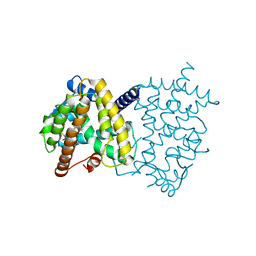 | | X-ray crystal structure of PPARgamma in complex with SR1643 | | Descriptor: | 4-bromo-N-{3,5-dichloro-4-[(quinolin-3-yl)oxy]phenyl}-2,5-difluorobenzene-1-sulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P, Kamenecka, T, Abell, A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPAR gamma-Targeted Antidiabetics.
J. Med. Chem., 60, 2017
|
|
5OY2
 
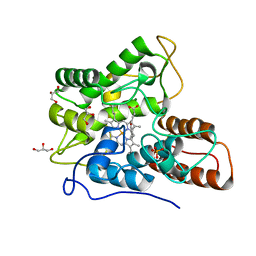 | | Direct-evolutioned unspecific peroxygenase from Agrocybe aegerita, in complex with DMP | | Descriptor: | 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2017-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Insights into the Substrate Promiscuity of a Laboratory-Evolved Peroxygenase.
Acs Chem.Biol., 13, 2018
|
|
4MJL
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and D-chiro-inositol | | Descriptor: | (1R,2R,3S,4S,5S,6S)-CYCLOHEXANE-1,2,3,4,5,6-HEXOL, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-03 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and D-chiro-inositol
To be Published
|
|
5OQG
 
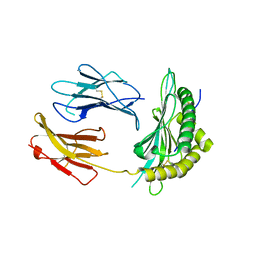 | |
3FWG
 
 | | Ferric camphor bound Cytochrome P450cam, Arg365Leu, Glu366Gln, monoclinic crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3WYC
 
 | | Structure of a meso-diaminopimelate dehydrogenase in complex with NADP | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ICC
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6N48
 
 | | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Liu, X, Masoudi, A, Kahsai, A.W, Huang, L.Y, Pani, B, Hirata, K, Ahn, S, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of beta2AR regulation by an intracellular positive allosteric modulator.
Science, 364, 2019
|
|
3ZCG
 
 | | Ascorbate peroxidase W41A-H42C mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASCORBATE PEROXIDASE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Probing the Conformational Mobility of the Active Site of a Heme Peroxidase.
Dalton Trans, 42, 2013
|
|
5UAM
 
 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Boniecki, M.T, Cygler, M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
4K02
 
 | | Crystal structure of AtDHNAT1, a 1,4-dihydroxy-2-naphthoyl-CoA thioesterase from Arabidopsis thaliana | | Descriptor: | 1,4-dihydroxy-2-naphthoyl-CoA thioesterase | | Authors: | Furt, F, Allen, W.J, Widhalm, J.R, Madzelan, P, Rizzo, R.C, Basset, G, Wilson, M.A. | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional convergence of structurally distinct thioesterases from cyanobacteria and plants involved in phylloquinone biosynthesis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3PRM
 
 | | Structural analysis of a viral OTU domain protease from the Crimean-Congo Hemorrhagic Fever virus in complex with human ubiquitin | | Descriptor: | Polyubiquitin-B (Fragment), RNA-directed RNA polymerase L | | Authors: | Capodagli, G.C, McKercher, M.A, Baker, E.A, Masters, E.M, Brunzelle, J.S, Pegan, S.D. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a viral ovarian tumor domain protease from the crimean-congo hemorrhagic Fever virus in complex with covalently bonded ubiquitin.
J.Virol., 85, 2011
|
|
3DFZ
 
 | | SirC, precorrin-2 dehydrogenase | | Descriptor: | GLYCEROL, Precorrin-2 dehydrogenase, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P, Warren, M.J. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase
Biochem.J., 415, 2008
|
|
3D8X
 
 | | Crystal Structure of Saccharomyces cerevisiae NDPPH Dependent Thioredoxin Reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Zhang, Z.Y, Bao, R, Yu, J, Chen, Y.X, Zhou, C.-Z. | | Deposit date: | 2008-05-26 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin
Biochim.Biophys.Acta, 1794, 2009
|
|
5UPX
 
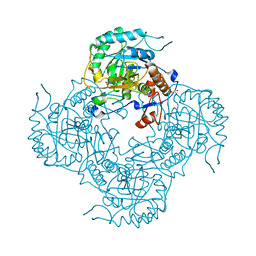 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the presence of Xanthosine Monophosphate | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Kim, Y, Makowska-Grzyska, M, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the presence of Xanthosine Monophosphate
To Be Published
|
|
1FC0
 
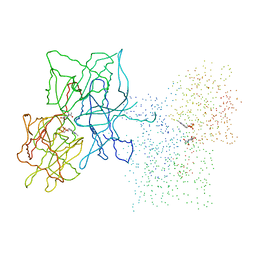 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE COMPLEXED WITH N-ACETYL-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | GLYCOGEN PHOSPHORYLASE, LIVER FORM, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.M, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
1T87
 
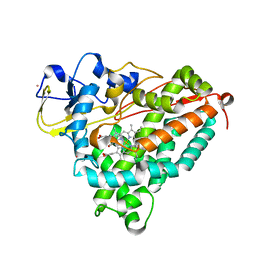 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CARBON MONOXIDE, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
3U4O
 
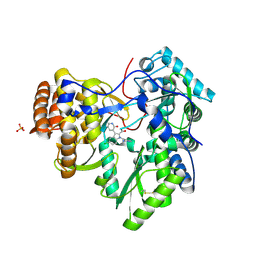 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1T88
 
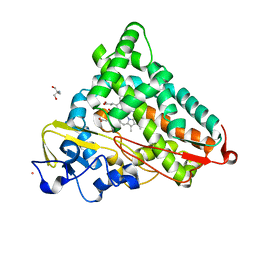 | | Crystal Structure of the Ferrous Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
3BLW
 
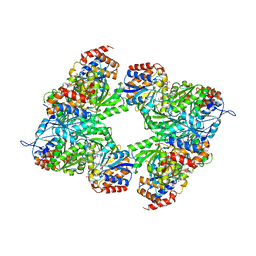 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
5OQH
 
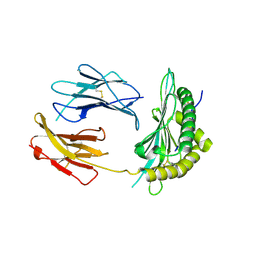 | | Crystal Structure of a disulfide trapped single chain trimer composed of the MHC I heavy chain H-2Kb Y84C K66A mutant, beta-2microglobulin, and ovalbumin-derived peptide | | Descriptor: | Beta-2-microglobulin,H-2 class I histocompatibility antigen, K-B alpha chain | | Authors: | Mikolajek, H, Werner, J.M, Beton, M.E. | | Deposit date: | 2017-08-11 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The partial dissociation of MHC class I-bound peptides exposes their N terminus to trimming by endoplasmic reticulum aminopeptidase 1.
J. Biol. Chem., 293, 2018
|
|
5OXU
 
 | |
1RW4
 
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
