6D3Z
 
 | | Protease SFTI complex | | Descriptor: | Plasminogen, Trypsin inhibitor 1 | | Authors: | Law, R.H.P, Wu, G. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Highly Potent and Selective Plasmin Inhibitors Based on the Sunflower Trypsin Inhibitor-1 Scaffold Attenuate Fibrinolysis in Plasma.
J. Med. Chem., 62, 2019
|
|
7OD1
 
 | |
5LOA
 
 | |
5LOL
 
 | | Glutathione-bound Dehydroascorbate Reductase 2 of Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase DHAR2, ... | | Authors: | Young, D.R, Pallo, A, Bodra, N, Messens, J. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arabidopsis thaliana dehydroascorbate reductase 2: Conformational flexibility during catalysis.
Sci Rep, 7, 2017
|
|
7OLS
 
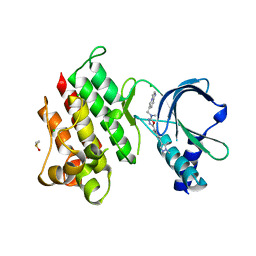 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
5LPF
 
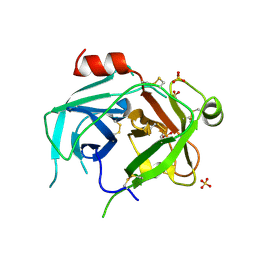 | | Kallikrein-related peptidase 10 | | Descriptor: | Kallikrein-10, SULFATE ION | | Authors: | Goettig, P, Debela, M, Magdolen, V, Bode, W, Brandstetter, H. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10.
Biol.Chem., 397, 2016
|
|
7OLV
 
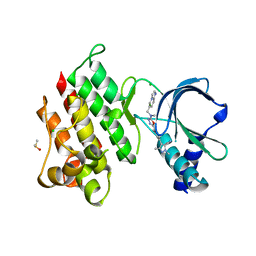 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl, cyano pyrazole group | | Descriptor: | 4-[4-[5-[imidazo[1,2-a]pyridin-6-ylmethyl(methyl)amino]-1,3,4-oxadiazol-2-yl]-3-methyl-phenyl]-2,5-dimethyl-pyrazole-3-carbonitrile, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
4W4N
 
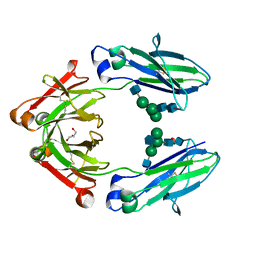 | | Crystal structure of human Fc at 1.80 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
4WE4
 
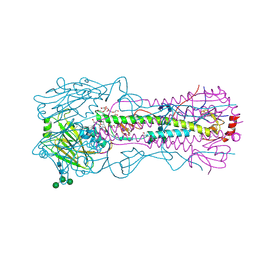 | | The crystal structure of hemagglutinin from 1968 H3N2 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
5LTM
 
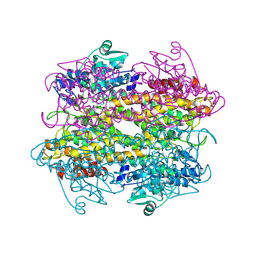 | |
6D25
 
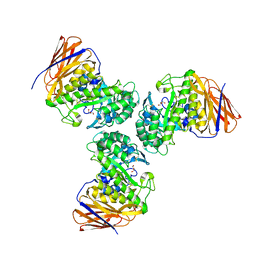 | | Crystal structure of the GH51 arabinofuranosidase from Xanthomonas axonopodis pv. citri | | Descriptor: | Alpha-L-arabinosidase, GLYCEROL | | Authors: | Santos, C.R, Morais, M.A.B, Tonoli, C.C.C, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-04-13 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The mechanism by which a distinguishing arabinofuranosidase can cope with internal di-substitutions in arabinoxylans.
Biotechnol Biofuels, 11, 2018
|
|
5LUG
 
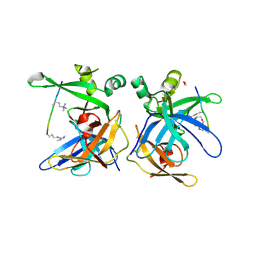 | | Crystal structure of human Spindlin-2B protein in complex with ART(M3L)QTA(2MR)KS peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-M3L-GLN-THR-ALA-2MR-LYS-SER, N3, ... | | Authors: | Srikannathasan, V, Gileadi, C, Talon, R, Shrestha, L, Kopec, J, Szykowska, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Oppermann, U, Huber, K. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Spindlin-2B protein in complex with ART(M3L)QTA(2MR)KS peptide
To be published
|
|
7OOI
 
 | | Anti-EphA1 JD1 VH domain | | Descriptor: | JD1 VH domain, SULFATE ION | | Authors: | Ereno-Orbea, J, Nilvebrant, J, Sidhu, S, Julien, J.P. | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains.
J.Mol.Biol., 433, 2021
|
|
5LYU
 
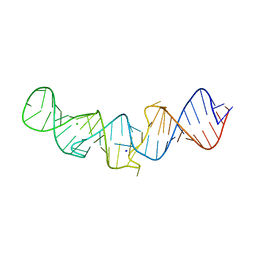 | | The native crystal structure of 7SK 5'-hairpin | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
5M2A
 
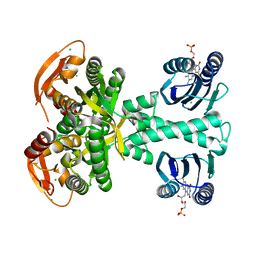 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-10-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
7O7G
 
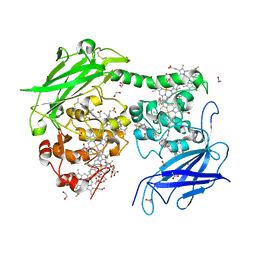 | | Crystal structure of the Shewanella oneidensis MR1 MtrC mutant H561M | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Edwards, M.J, van Wonderen, J.H, Newton-Payne, S.E, Butt, J.N, Clarke, T.A. | | Deposit date: | 2021-04-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanosecond heme-to-heme electron transfer rates in a multiheme cytochrome nanowire reported by a spectrally unique His/Met-ligated heme.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OJY
 
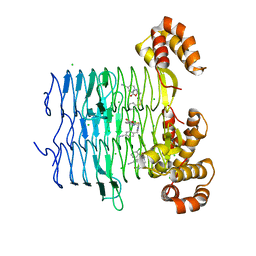 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 6 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-oxidanyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OKB
 
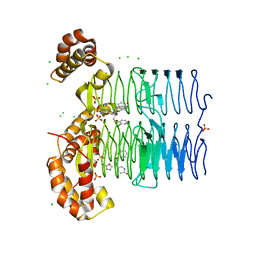 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 45 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OKC
 
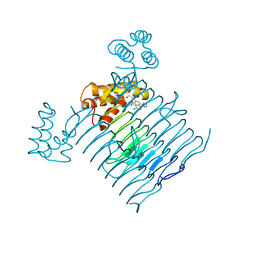 | | Crystal structure of Escherichia coli LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SODIUM ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7O66
 
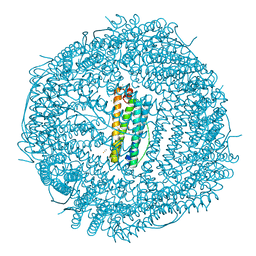 | | Crystal structure of human mitochondrial ferritin (hMTF) Fe(II)-loaded for 60 minutes showing either a dioxygen or a superoxide anion coordinated to iron ions in the ferroxidase site | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Ciambellotti, S, Tassone, G, Turano, P, Mangani, S. | | Deposit date: | 2021-04-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Iron Binding in the Ferroxidase Site of Human Mitochondrial Ferritin.
Chemistry, 27, 2021
|
|
7O65
 
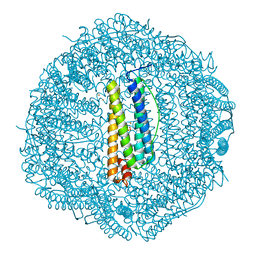 | | Crystal structure of human mitochondrial ferritin (hMTF) Fe(II)-loaded for 90 minutes showing either a dioxygen or a superoxide anion coordinated to iron ions in the ferroxidase site | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Ciambellotti, S, Tassone, G, Turano, P, Mangani, S. | | Deposit date: | 2021-04-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron Binding in the Ferroxidase Site of Human Mitochondrial Ferritin.
Chemistry, 27, 2021
|
|
4URR
 
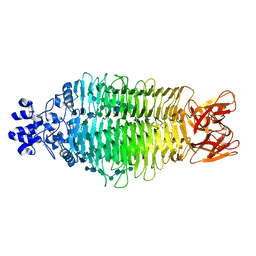 | | Tailspike protein of Sf6 bacteriophage bound to Shigella flexneri O- antigen octasaccharide fragment | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL TAIL PROTEIN, MANGANESE (II) ION, ... | | Authors: | Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2014-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein-Polysaccharide Interactions.
J.Am.Chem.Soc., 138, 2016
|
|
7OMN
 
 | | Anti-EphA1 JD1-1 VH domain | | Descriptor: | GLYCEROL, IMIDAZOLE, JD1-1 VH domain | | Authors: | Ereno Orbea, J, Nilvebrant, J, Sidhu, S, Julien, J.P. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains.
J.Mol.Biol., 433, 2021
|
|
6CEW
 
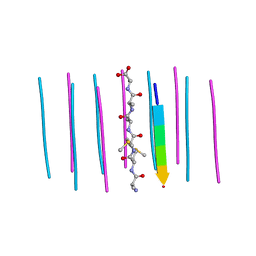 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
