9E1H
 
 | |
9E17
 
 | | Structure of RyR1 in the primed state in the presence of caffeine (reprocessed/reanalyzed from EMPIAR-10997, 7TZC, EMD-26205) | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2024-10-21 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Targeting Ryanodine Receptors with Allopurinol and Xanthine Derivatives for the Treatment of Cardiac and Musculoskeletal Weakness Disorders
To Be Published
|
|
9E1G
 
 | |
9E1I
 
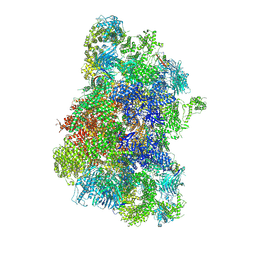 | | Structure of RyR1 in the open state in the presence of oxopyricid | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2024-10-21 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Targeting Ryanodine Receptors with Allopurinol and Xanthine Derivatives for the Treatment of Cardiac and Musculoskeletal Weakness Disorders
To Be Published
|
|
2IF4
 
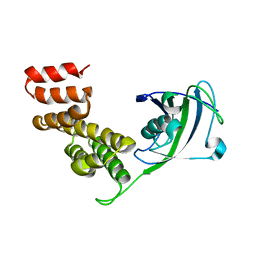 | |
8RRW
 
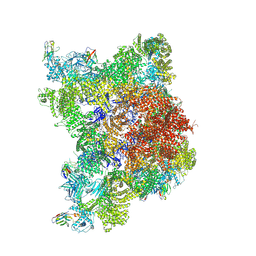 | |
8RRV
 
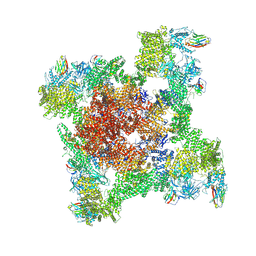 | |
8RRT
 
 | |
8RS0
 
 | |
8RRU
 
 | |
8RRX
 
 | | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8SER
 
 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEP
 
 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SES
 
 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
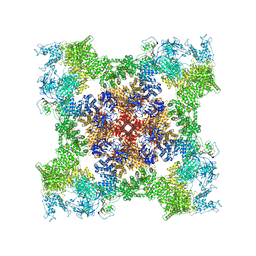 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
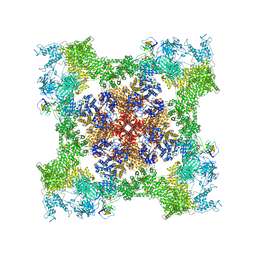 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
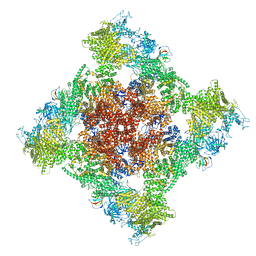 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
1P5Q
 
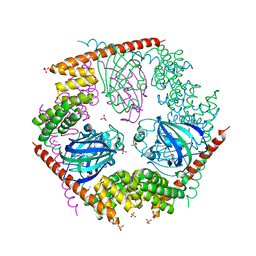 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q6U
 
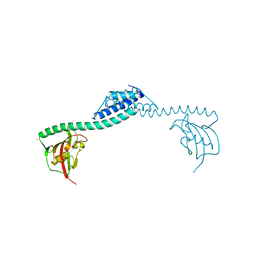 | | Crystal structure of FkpA from Escherichia coli | | Descriptor: | CESIUM ION, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-14 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6I
 
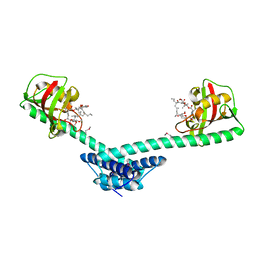 | | Crystal structure of a truncated form of FkpA from Escherichia coli, in complex with immunosuppressant FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6H
 
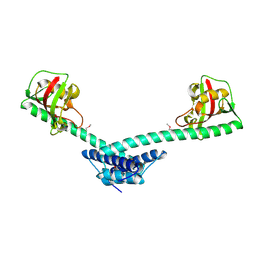 | | Crystal structure of a truncated form of FkpA from Escherichia coli | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1QZ2
 
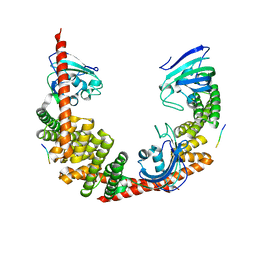 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1NSG
 
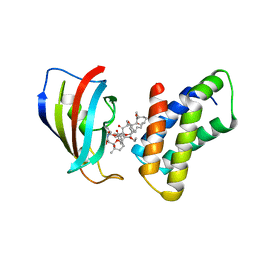 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FKBP-RAPAMYCIN ASSOCIATED PROTEIN (FRAP) | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8UQ3
 
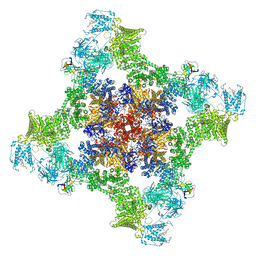 | | Structure of human RyR2-S2808D in the closed state in the presence of ARM210 | | Descriptor: | 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2023-10-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for ryanodine receptor type 2 leak in heart failure and arrhythmogenic disorders.
Nat Commun, 15, 2024
|
|
