4MHE
 
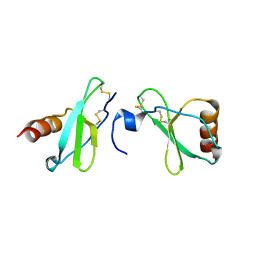 | | Crystal structure of CC-chemokine 18 | | Descriptor: | ACETATE ION, C-C motif chemokine 18 | | Authors: | Liang, W.G, Tang, W.-J. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4D7G
 
 | | Human FXIa in complex with small molecule inhibitors. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COAGULATION FACTOR XIA, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Jacso, T, Ullah, V, Redzick, A, Borjesson, U, Olsson, T, Norberg, M, Akerud, T. | | Deposit date: | 2014-11-24 | | Release date: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | From Mm Fragments to Nm Compounds Using Iloe-NMR to Guide Linking of Compounds in Fragment Based Drug Discovery (Fbdd).
To be Published
|
|
4D83
 
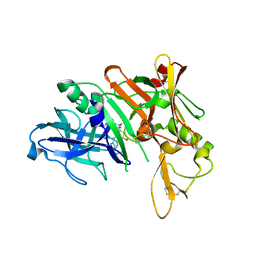 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BUR436, derived from a co-crystallization experiment | | Descriptor: | (3R,4S,5S)-3-[(3-tert-butylbenzyl)amino]-5-{[3-(2,2-difluoroethyl)-1H-indol-5-yl]methyl}tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
5LEF
 
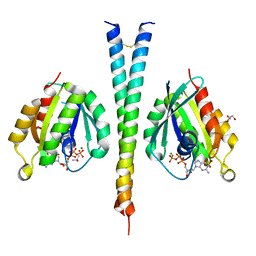 | | Rab6A:Kif20A complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ISOPROPYL ALCOHOL, ... | | Authors: | Bressanelli, G, Pylypenko, O, Houdusse, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Coupling fission and exit of RAB6 vesicles at Golgi hotspots through kinesin-myosin interactions.
Nat Commun, 8, 2017
|
|
4D8N
 
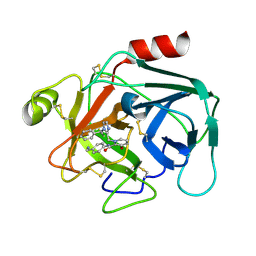 | |
4LOP
 
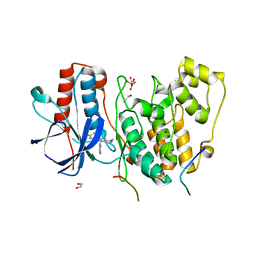 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4D9U
 
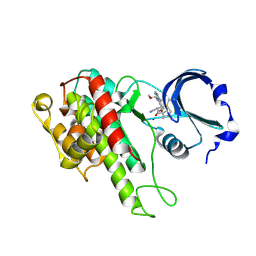 | | Rsk2 C-terminal Kinase Domain, (E)-tert-butyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, tert-butyl (2S)-3-[4-amino-7-(3-hydroxypropyl)-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
4D9Z
 
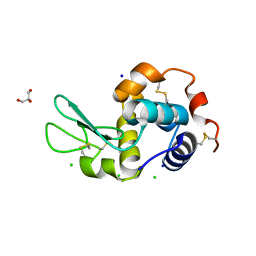 | | Lysozyme at 318K | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
4CRF
 
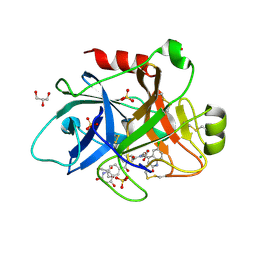 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | CHLORIDE ION, Coagulation factor XIa light chain, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
2W1M
 
 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 2.070 A WAVELENGTH with 2theta 30 degrees data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VNT
 
 | | UROKINASE-TYPE PLASMINOGEN ACTIVATOR INHIBITOR COMPLEX WITH A 1-(7- SULPHOAMIDOISOQUINOLINYL)GUANIDINE | | Descriptor: | 1-({4-CHLORO-1-[(DIAMINOMETHYLIDENE)AMINO]ISOQUINOLIN-7-YL}SULFONYL)-D-PROLINE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Fish, P.V, Barber, C.G, Brown, D.G, Butt, R, Henry, B.T, Horne, V, Huggins, J.P, Mccleverty, D, Phillips, C, Webster, R, Dickinson, R.P, Collis, M.G, King, E, O'Gara, M, Mcintosh, F. | | Deposit date: | 2008-02-07 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Urokinase-Type Plasminogen Activator (Upa) Inhibitors 4. 1-(7-Sulphonamidoisoquinolinyl) Guanidines
J.Med.Chem., 50, 2007
|
|
2WAR
 
 | | Hen Egg White Lysozyme E35Q chitopentaose complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME C | | Authors: | Davies, G.J, Withers, S.G, Vocadlo, D.J. | | Deposit date: | 2009-02-13 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Chitopentaose Complex of a Mutant Hen Egg-White Lysozyme Displays No Distortion of the -1 Sugar Away from a 4C1 Chair Conformation
Aust.J.Chem., 62, 2009
|
|
5LHP
 
 | | The p-aminobenzamidine active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment, P-AMINO BENZAMIDINE, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
8HGZ
 
 | | Crystal structure of insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | DeMirci, H, Ayan, E. | | Deposit date: | 2022-11-16 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative Study of High-Resolution LysB29(N epsilon-myristoyl) des(B30) Insulin Structures Display Novel Dynamic Causal Interrelations in Monomeric-Dimeric Motions
Crystals, 13, 2023
|
|
2VUZ
 
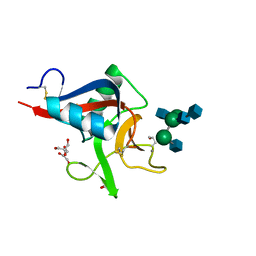 | | Crystal structure of Codakine in complex with biantennary nonasaccharide at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
2W1Y
 
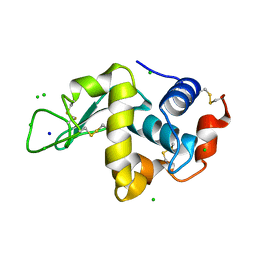 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 1.540 A wavelength 180 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VNW
 
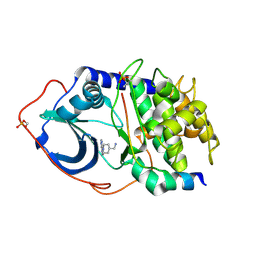 | | Structure of PKA-PKB chimera complexed with (1-(9H-Purin-6-yl) piperidin-4-yl)methanamine | | Descriptor: | 1-[1-(9H-purin-6-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
5LPV
 
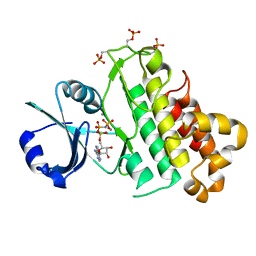 | |
2VYZ
 
 | | Mutant Ala55Phe of Cerebratulus lacteus mini-hemoglobin | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
4M3M
 
 | |
2WF3
 
 | | Human BACE-1 in complex with 6-(ethylamino)-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-1, 3,4,5-tetrahydro-2,1-benzothiazepine-8-carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, GLYCEROL, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-6-(ETHYLAMINO)-1-METHYL-1,3,4,5-TETRAHYDRO-2,1-BENZOTHIAZEPINE-8-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4MBI
 
 | | Discovery of Pyrazolo[1,5a]pyrimidine-based Pim1 Inhibitors | | Descriptor: | N,N-dimethyl-N'-[3-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]ethane-1,2-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Azevedo, R, Fischmann, T.O. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2VO3
 
 | | Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine | | Descriptor: | 1-[4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
4M4F
 
 | | Radiation damage study of Cu T6-insulin - 0.01 MGy | | Descriptor: | COPPER (II) ION, Insulin | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M4M
 
 | | The structure of Ni T6 bovine insulin | | Descriptor: | Insulin, NICKEL (II) ION | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
