7CMJ
 
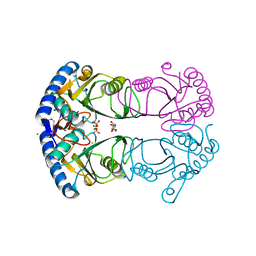 | |
7DNF
 
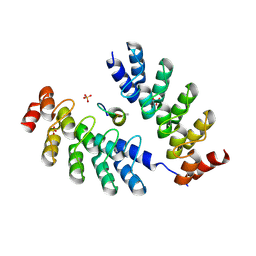 | | DARPin 63_B7 in complex with V3-IY (MN) crown mimetic | | Descriptor: | DARPin 63_B7, SULFATE ION, V3-IY (MN) crown mimetic peptide | | Authors: | Wu, Y, Plueckthun, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7DNG
 
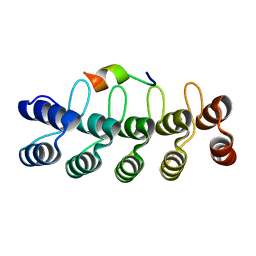 | |
7DNE
 
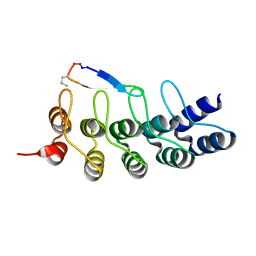 | |
7CCN
 
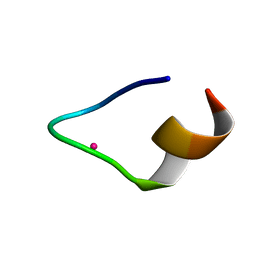 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | Descriptor: | LBT3, LUTETIUM (III) ION | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7E6G
 
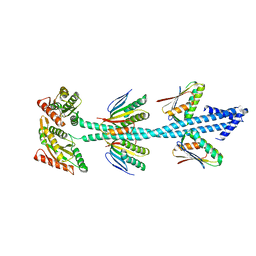 | | Crystal structure of diguanylate cyclase SiaD in complex with its activator SiaC from Pseudomonas aeruginosa | | Descriptor: | DUF1987 domain-containing protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa .
Elife, 10, 2021
|
|
7C0N
 
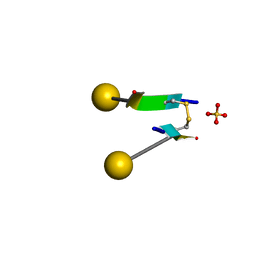 | | Crystal structure of a self-assembling galactosylated peptide homodimer | | Descriptor: | SULFATE ION, Self-assembling galactosylated tyrosine-rich peptide, beta-D-galactopyranose | | Authors: | He, C, Wu, S, Chi, C, Zhang, W, Ma, M, Lai, L, Dong, S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Glycopeptide Self-Assembly Modulated by Glycan Stereochemistry through Glycan-Aromatic Interactions.
J.Am.Chem.Soc., 142, 2020
|
|
7BID
 
 | | Crystal structure of v31WRAP-T, a 7-bladed designer protein | | Descriptor: | v31WRAP-T | | Authors: | Laier, I, Mylemans, B, Lee, X.Y, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIF
 
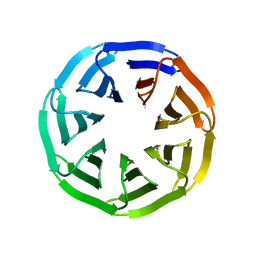 | | Crystal structure of v22WRAP-T, a 7-bladed designer protein | | Descriptor: | v22WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIG
 
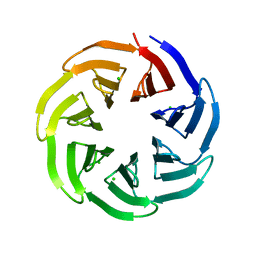 | | Crystal structure of v13WRAP-T, a 7-bladed designer protein | | Descriptor: | CHLORIDE ION, v13WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIE
 
 | | Crystal structure of nvWrap-T, a 7-bladed symmetric propeller | | Descriptor: | CITRIC ACID, nvWRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7EQ9
 
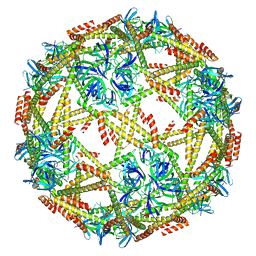 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | Descriptor: | TIP60 | | Authors: | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
7F1I
 
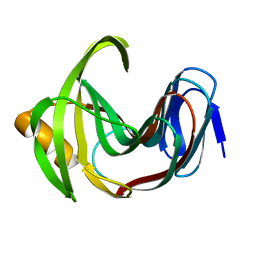 | | Designed enzyme RA61 M48K/I72D mutant: form II | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1H
 
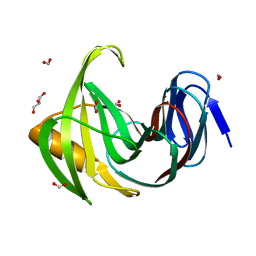 | | Designed enzyme RA61 M48K/I72D mutant: form I | | Descriptor: | Engineered Retroaldolase, FORMIC ACID, GLYCEROL | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1J
 
 | | Designed enzyme RA61 M48K/I72D mutant: form III | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1K
 
 | | Designed enzyme RA61 M48K/I72D mutant: form IV | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | Descriptor: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
5H1H
 
 | | NMR structure of SLBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
5H1I
 
 | | NMR structure of TIBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
6JOP
 
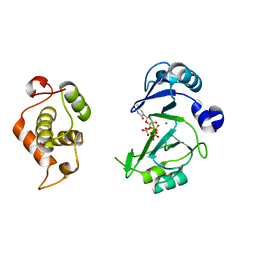 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | MAGNESIUM ION, Primase, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
5H78
 
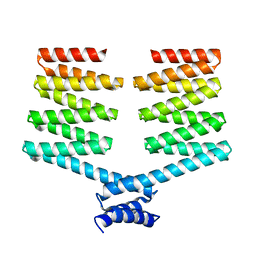 | | Crystal structure of the PKA-DHR14 fusion protein | | Descriptor: | cAMP-dependent protein kinase type II-alpha regulatory subunit,DHR14 | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5HPP
 
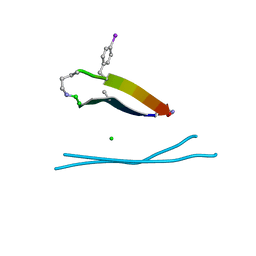 | |
6JOQ
 
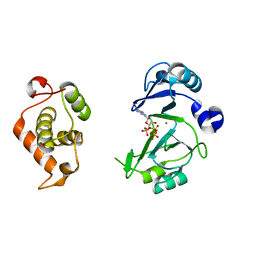 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
5H7C
 
 | | Crystal structure of a repeat protein with two Protein A-DHR14 repeat modules | | Descriptor: | Immunoglobulin G-binding protein A, DHR14 | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5HI1
 
 | | Backbone Modifications in the Protein GB1 Helix: Aib24, beta-3-Lys28, beta-3-Lys31, Aib35 | | Descriptor: | ACETATE ION, Immunoglobulin G-binding protein G | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
