1Y0V
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and pyrophosphate | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.-J. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor
Embo J., 24, 2005
|
|
1Y0K
 
 | | Structure of Protein of Unknown Function PA4535 from Pseudomonas aeruginosa strain PAO1, Monooxygenase Superfamily | | Descriptor: | hypothetical protein PA4535 | | Authors: | Nocek, B.P, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 A Crystal Structure of the Hypothetical Protein Pa4535 from Pseudomonas Aeruginosa
To be Published
|
|
1YC2
 
 | | Sir2Af2-NAD-ADPribose-nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase 2, ... | | Authors: | Avalos, J.L, Bever, M.K, Wolberger, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD(+) Cosubstrate Specificity of a Sir2 Enzyme.
Mol.Cell, 17, 2005
|
|
1Y9F
 
 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-allyl Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2AT)P*AP*CP*GP*C)-3') | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-15 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
1YE9
 
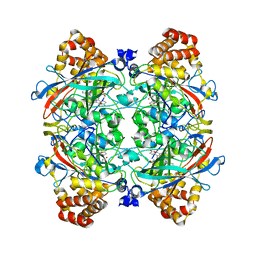 | | Crystal structure of proteolytically truncated catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, catalase HPII | | Authors: | Loewen, P.C, Chelikani, P, Carpena, X, Fita, I, Perez-Luque, R, Donald, L.J, Switala, J, Duckworth, H.W. | | Deposit date: | 2004-12-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a Large Subunit Catalase Truncated by Proteolytic Cleavage(,)
Biochemistry, 44, 2005
|
|
1YG6
 
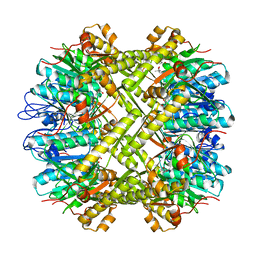 | | ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Bewley, M.C, Graziano, V, Griffin, K, Flanagan, J.M. | | Deposit date: | 2005-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes.
J.Struct.Biol., 153, 2006
|
|
2AAM
 
 | |
1YRQ
 
 | | Structure of the ready oxidized form of [NiFe]-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
1YQ5
 
 | | PRD1 vertex protein P5 | | Descriptor: | Minor capsid protein | | Authors: | Merckel, M.C, Huiskonen, J.T, Goldman, A, Bamford, D.H, Tuma, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture.
Mol.Cell, 18, 2005
|
|
2C9B
 
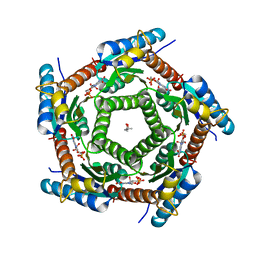 | | Lumazine Synthase from Mycobacterium tuberculosus Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL), 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
1YQW
 
 | | Structure of the Oxidized Unready Form of Ni-Fe Hydrogenase | | Descriptor: | BICARBONATE ION, CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, ... | | Authors: | Volbeda, A. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
2CE8
 
 | |
2BAZ
 
 | |
2CJF
 
 | | TYPE II DEHYDROQUINASE INHIBITOR COMPLEX | | Descriptor: | (1S,4S,5S)-1,4,5-TRIHYDROXY-3-[3-(PHENYLTHIO)PHENYL]CYCLOHEX-2-ENE-1-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Payne, R.J, Riboldi-Tunnicliffe, A, Abell, A.D, Lapthorn, A.J, Abell, C. | | Deposit date: | 2006-03-31 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, and structural studies on potent biaryl inhibitors of type II dehydroquinases.
Chemmedchem, 2, 2007
|
|
2C91
 
 | | mouse succinic semialdehyde reductase, AKR7A5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AFLATOXIN B1 ALDEHYDE REDUCTASE MEMBER 2, GLYCEROL, ... | | Authors: | Zhu, X, Ellis, E.M, Lapthorn, A.J. | | Deposit date: | 2005-12-08 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mouse Succinic Semialdehyde Reductase Akr7A5: Structural Basis for Substrate Specificity.
Biochemistry, 45, 2006
|
|
2CA9
 
 | | apo-NIKR from helicobacter pylori in closed trans-conformation | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2CAD
 
 | | NikR from Helicobacter pylori in closed trans-conformation and nickel bound to 2F, 2X and 2I sites. | | Descriptor: | CITRIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2CJT
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
2D8P
 
 | | Structure of HYPER-VIL-thaumatin | | Descriptor: | IODIDE ION, Thaumatin I | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D90
 
 | |
1TUO
 
 | | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8 | | Descriptor: | Putative phosphomannomutase | | Authors: | Misaki, S, Suzuki, S, Fujimoto, S, Sakurai, M, Kobayashi, M, Nishijima, K, Kunishima, N, Sugawara, M, Kuroishi, C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1TV7
 
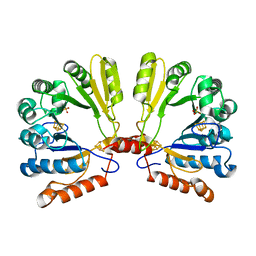 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1U6S
 
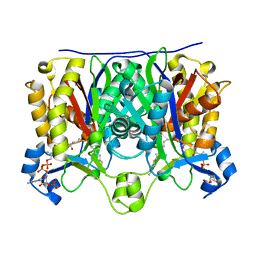 | | Crystal Structure of the Complex Between Mycobacterium Tuberculosis Beta-Ketoacyl-Acyl Carrier Protein Synthase III and Lauroyl Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III, DODECYL-COA | | Authors: | Musayev, F, Sachdeva, S, Scarsdale, J.N, Reynolds, K.A, Wright, H.T. | | Deposit date: | 2004-07-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A.
J.Mol.Biol., 346, 2005
|
|
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
1U6I
 
 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
