2FDC
 
 | |
2FDD
 
 | | Crystal structure of HIV protease D545701 bound with GW0385 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-3-[(1,3-BENZODIOXOL-5-YLSULFONYL)(ISOBUTYL)AMINO]-2-HYDROXY-1-{4-[(2-METHYL-1,3-THIAZOL-4-YL)METHOXY]BENZYL}PROPYL]CARBAMATE, Gag-Pol polyprotein | | Authors: | Xu, R.X. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: The discovery of GW0385.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FDE
 
 | | Wild type HIV protease bound with GW0385 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-3-[(1,3-BENZODIOXOL-5-YLSULFONYL)(ISOBUTYL)AMINO]-2-HYDROXY-1-{4-[(2-METHYL-1,3-THIAZOL-4-YL)METHOXY]BENZYL}PROPYL]CARBAMATE, Gag-Pol polyprotein, POTASSIUM ION | | Authors: | Xu, R.X. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: The discovery of GW0385.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FDF
 
 | | Crystal Structure of AlkB in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDG
 
 | | Crystal Structure of AlkB in complex with Fe(II), succinate, and methylated trinucleotide T-meA-T | | Descriptor: | 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, FE (II) ION, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDH
 
 | | Crystal Structure of AlkB in complex with Mn(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDI
 
 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T (air 3 hours) | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDJ
 
 | | Crystal Structure of AlkB in complex with Fe(II) and succinate | | Descriptor: | Alkylated DNA repair protein alkB, FE (II) ION, SUCCINIC ACID | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDK
 
 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T (air 9 days) | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDM
 
 | | Crystal structure of the ternary complex of signalling glycoprotein frm sheep (SPS-40)with hexasaccharide (NAG6) and peptide Trp-Pro-Trp at 3.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Somvanshi, R.K, Sharma, S, Singh, T.P. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the ternary complex of signalling glycoprotein frm sheep (SPS-40)
with hexasaccharide (NAG6) and peptide Trp-Pro-Trp at 3.0A resolution
To be Published
|
|
2FDN
 
 | | 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDI-URICI | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Meyer, J, Moulis, J.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic resolution (0.94 A) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein.
Biochemistry, 36, 1997
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
2FDQ
 
 | |
2FDR
 
 | | Crystal Structure of Conserved Haloacid Dehalogenase-like Protein of Unknown Function ATU0790 from Agrobacterium tumefaciens str. C58 | | Descriptor: | MAGNESIUM ION, conserved hypothetical protein | | Authors: | Nocek, B, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hydrolases/phosphatases-like fold protein
from Agrobacterium tumefaciens str. C58
To be Published
|
|
2FDS
 
 | | Crystal Structure of Plasmodium Berghei Orotidine 5'-monophosphate Decarboxylase (ortholog of Plasmodium falciparum PF10_0225) | | Descriptor: | IODIDE ION, orotidine-monophosphate-decarboxylase | | Authors: | Qiu, W, Dong, A, Wasney, G, Vedadi, M, Lew, J, Kozieradski, I, Alam, Z, Melone, M, Weigelt, J, Sundstrom, M, Edwards, A, Arrowsmith, C, Hui, R, Gao, M, Bochkarev, A, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2FDT
 
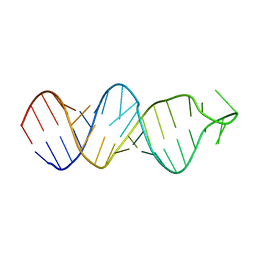 | | Solution structure of a conserved RNA hairpin of eel LINE UnaL2 | | Descriptor: | 36-mer | | Authors: | Nomura, Y, Baba, S, Nakazato, S, Sakamoto, T, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2
Nucleic Acids Res., 34, 2006
|
|
2FDU
 
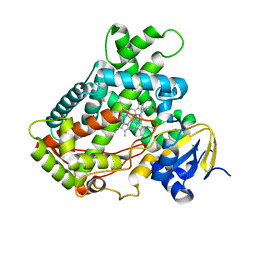 | | Microsomal P450 2A6 with the inhibitor N,N-Dimethyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | Cytochrome P450 2A6, N,N-DIMETHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FDV
 
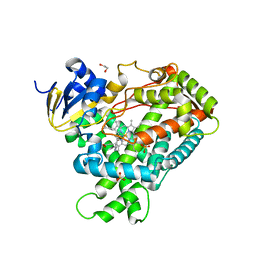 | | Microsomal P450 2A6 with the inhibitor N-Methyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 2A6, N-METHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FDW
 
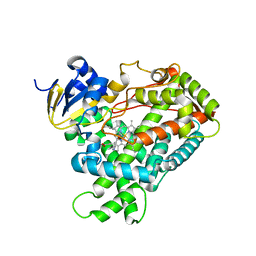 | | Crystal Structure Of Human Microsomal P450 2A6 with the inhibitor (5-(Pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | (5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FDX
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT N137A OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
2FDY
 
 | | Microsomal P450 2A6 with the inhibitor Adrithiol bound | | Descriptor: | 4,4'-DIPYRIDYL DISULFIDE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FE0
 
 | |
2FE1
 
 | | Crystal Structure of PAE0151 from Pyrobaculum aerophilum | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bunker, R.D, Baker, E.N, Arcus, V.L. | | Deposit date: | 2005-12-15 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon.
Proteins, 72, 2008
|
|
2FE3
 
 | |
