2F71
 
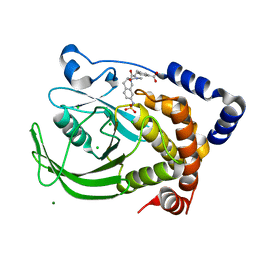 | | Protein tyrosine phosphatase 1B with sulfamic acid inhibitors | | Descriptor: | 3-[3-(3(S)-METHYLCARBAMOYL-7-SULFOAMINO-3,4,-DIHYDRO-1H-ISOQUINOLIN-2-YL)-3-OXO-PROPYL]-BENZOIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Klopfenstein, S.R. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1,2,3,4-Tetrahydroisoquinolinyl sulfamic acids as phosphatase PTP1B inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F73
 
 | | Crystal structure of human fatty acid binding protein 1 (FABP1) | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Kursula, P, Thorsell, A.G, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, van den Berg, S, Weigelt, J, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human FABP1
To be Published
|
|
2F74
 
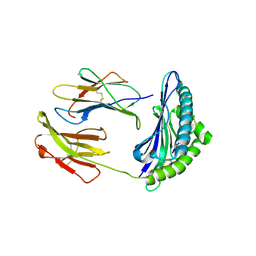 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
2F76
 
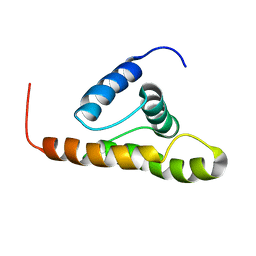 | | Solution structure of the M-PMV wild type matrix protein (p10) | | Descriptor: | Core protein p10 | | Authors: | Vlach, J, Lipov, J, Veverka, V, Lang, J, Srb, P, Rumlova, M, Hunter, E, Ruml, T, Hrabal, R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2F77
 
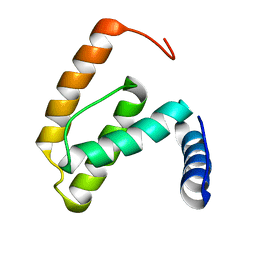 | | Solution structure of the R55F mutant of M-PMV matrix protein (p10) | | Descriptor: | Core protein p10 | | Authors: | Vlach, J, Lipov, J, Veverka, V, Lang, J, Srb, P, Rumlova, M, Hunter, E, Ruml, T, Hrabal, R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2F78
 
 | | BenM effector binding domain with its effector benzoate | | Descriptor: | BENZOIC ACID, HTH-type transcriptional regulator benM | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F7A
 
 | | BenM effector binding domain with its effector, cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, ACETATE ION, BENZOIC ACID, ... | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F7B
 
 | | CatM effector binding domain | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator catM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F7C
 
 | | CatM effector binding domain with its effector cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, HTH-type transcriptional regulator catM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F7D
 
 | | A mutant rabbit cathepsin K with a nitrile inhibitor | | Descriptor: | (1R,2R)-N-(2-AMINOETHYL)-2-{[(4-METHOXYPHENYL)SULFONYL]METHYL}CYCLOHEXANECARBOXAMIDE, Cathepsin K | | Authors: | Somoza, J.R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-03-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Beta-substituted cyclohexanecarboxamide: a nonpeptidic framework for the design of potent inhibitors of cathepsin K.
J.Med.Chem., 49, 2006
|
|
2F7E
 
 | | PKA complexed with (S)-2-(1H-Indol-3-yl)-1-(5-isoquinolin-6-yl-pyridin-3-yloxymethyl-etylamine | | Descriptor: | (1S)-2-(1H-INDOL-3-YL)-1-{[(5-ISOQUINOLIN-6-YLPYRIDIN-3-YL)OXY]METHYL}ETHYLAMINE, PKI inhibitory peptide, cAMP-dependent protein kinase, ... | | Authors: | Stoll, V.S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F7F
 
 | | Crystal structure of Enterococcus faecalis putative nicotinate phosphoribosyltransferase, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | DIPHOSPHATE, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enterococcus Faecalis Nicotinate Phosphoribosyltransferase
To be Published
|
|
2F7I
 
 | | Human transthyretin (TTR) complexed with diflunisal analogues- TTR. 2',6'-Difluorobiphenyl-4-carboxylic Acid | | Descriptor: | 2',6'-DIFLUOROBIPHENYL-4-CARBOXYLIC ACID, Transthyretin | | Authors: | Palaninathan, S.K, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diflunisal Analogues Stabilize the Native State of Transthyretin. Potent Inhibition of Amyloidogenesis.
J.Med.Chem., 47, 2004
|
|
2F7K
 
 | |
2F7L
 
 | | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase | | Descriptor: | 455aa long hypothetical phospho-sugar mutase, SULFATE ION | | Authors: | Kawamura, T, Sakai, N, Akutsu, J, Zhang, Z, Watanabe, N, Kawarabayashi, Y, Tanaka, I. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii phosphomannomutase/phosphoglucomutase
To be Published
|
|
2F7M
 
 | | Crystal Structure of Unliganded Human FPPS | | Descriptor: | Farnesyl Diphosphate Synthase, PHOSPHATE ION | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2F7N
 
 | | Structure of D. radiodurans Dps-1 | | Descriptor: | COBALT (II) ION, DNA-binding stress response protein, Dps family, ... | | Authors: | Lee, Y.H, Kim, S.G, Bhattacharyya, G, Grove, A. | | Deposit date: | 2005-12-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans.
J.Mol.Biol., 361, 2006
|
|
2F7O
 
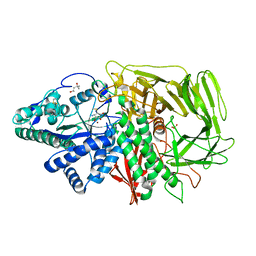 | | Golgi alpha-mannosidase II complex with mannostatin A | | Descriptor: | (1R,2R,3R,4S,5R)-4-AMINO-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis of the Inhibition of Golgi alpha-Mannosidase II by Mannostatin A and the Role of the Thiomethyl Moiety in Ligand-Protein Interactions.
J.Am.Chem.Soc., 128, 2006
|
|
2F7P
 
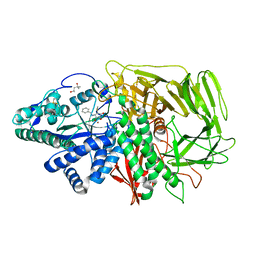 | | Golgi alpha-mannosidase II complex with benzyl-mannostatin A | | Descriptor: | (1R,2R,3R,4S,5R)-4-(BENZYLAMINO)-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Basis of the Inhibition of Golgi alpha-Mannosidase II by Mannostatin A and the Role of the Thiomethyl Moiety in Ligand-Protein Interactions.
J.Am.Chem.Soc., 128, 2006
|
|
2F7Q
 
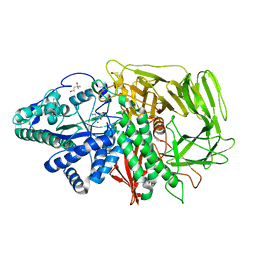 | | Golgi alpha-mannosidase II complex with aminocyclopentitetrol | | Descriptor: | (1R,2R,3S,4S,5R)-5-AMINOCYCLOPENTANE-1,2,3,4-TETROL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Golgi alpha-mannosidase II complex with aminocyclopentitetrol
To be Published
|
|
2F7R
 
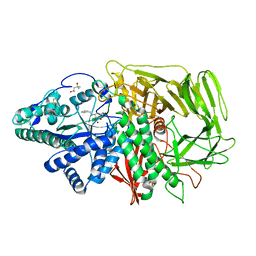 | | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol | | Descriptor: | (1R,2R,3S,4S,5R)-5-(BENZYLAMINO)CYCLOPENTANE-1,2,3,4-TETROL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol
To be Published
|
|
2F7S
 
 | | The crystal structure of human Rab27b bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Wang, J, Ismail, S, Shen, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of human Rab27b
To be Published
|
|
2F7T
 
 | | Crystal structure of the catalytic domain of Mos1 mariner transposase | | Descriptor: | MAGNESIUM ION, Mos1 transposase | | Authors: | Richardson, J.M, Dawson, A, Taylor, P, Finnegan, D.J, Walkinshaw, M.D. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Mos1 transposition: insights from structural analysis
Embo J., 25, 2006
|
|
2F7V
 
 | | Structure of acetylcitrulline deacetylase complexed with one Co | | Descriptor: | COBALT (II) ION, aectylcitrulline deacetylase | | Authors: | Shi, D, Yu, X, Roth, L, Allewell, N.M, Tuchman, M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a novel N-acetyl-L-citrulline deacetylase from Xanthomonas campestris
Biophys.Chem., 126, 2007
|
|
2F7W
 
 | |
