3DZP
 
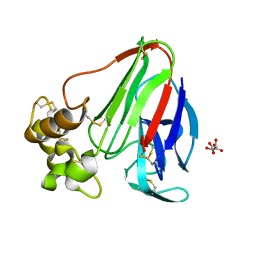 | |
1GZ0
 
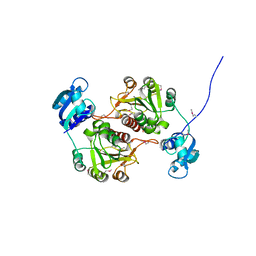 | | 23S RIBOSOMAL RNA G2251 2'O-METHYLTRANSFERASE RLMB | | Descriptor: | HYPOTHETICAL TRNA/RRNA METHYLTRANSFERASE YJFH | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-05-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Rlmb 23S Rrna Methyltransferase Reveals a New Methyltransferase Fold with a Unique Knot
Structure, 10, 2002
|
|
2I3V
 
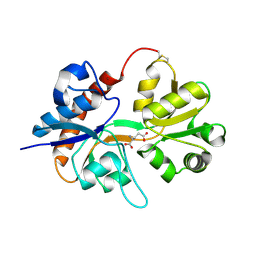 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
2CIQ
 
 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HEXOSE-6-PHOSPHATE MUTAROTASE, ... | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
3DZN
 
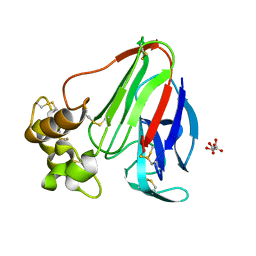 | |
1Y0S
 
 | | Crystal structure of PPAR delta complexed with GW2331 | | Descriptor: | (2S)-2-(4-[2-(3-[2,4-DIFLUOROPHENYL]-1-HEPTYLUREIDO)ETHYL]PHENOXY)-2-METHYLBUTYRIC ACID, IODIDE ION, Peroxisome proliferator activated receptor delta, ... | | Authors: | Takada, I, Yu, R.T, Xu, H.E, Xu, R.X, Lambert, M.H, Montana, V.G, Kliewer, S.A, Evans, R.M, Umesono, K. | | Deposit date: | 2004-11-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Alteration of a Single Amino Acid in Peroxisome Proliferator-Activated Receptor-alpha (PPARalpha) Generates a PPAR delta Phenotype
MOL.ENDOCRINOL., 14, 2000
|
|
2I3W
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
7C10
 
 | | Dithiol cGrx1 | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1ZTU
 
 | | Structure of the chromophore binding domain of bacterial phytochrome | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Forest, K.T, Vierstra, R.D. | | Deposit date: | 2005-05-27 | | Release date: | 2005-11-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome.
Nature, 438, 2005
|
|
2OBU
 
 | |
1MR0
 
 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
2BOU
 
 | | EGF Domains 1,2,5 of human EMR2, a 7-TM immune system molecule, in complex with barium. | | Descriptor: | BARIUM ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Abbott, R.J.M, Spendlove, I, Roversi, P, Teriete, P, Knott, V, Handford, P.A, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2005-04-14 | | Release date: | 2006-10-18 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of a Novel T Cell Receptor Co-Regulatory Protein Complex, Cd97-Cd55.
J.Biol.Chem., 282, 2007
|
|
2LX3
 
 | | 1H,13C,15N assignments for an isoform of the type III antifreeze protein from notched-fin eelpout | | Descriptor: | Type III antifreeze protein nfeAFP11 | | Authors: | Kumeta, H, Ogura, K, Nishimiya, Y, Miura, A, Inagaki, F, Tsuda, S. | | Deposit date: | 2012-08-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: a defective isoform and its activity-improved variant of a type III antifreeze protein from Zoarces elongates Kner
J.Biomol.Nmr, 55, 2013
|
|
1EMN
 
 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
1EMO
 
 | | NMR STUDY OF A PAIR OF FIBRILLIN CA2+ BINDING EPIDERMAL GROWTH FACTOR-LIKE DOMAINS, 22 STRUCTURES | | Descriptor: | CALCIUM ION, FIBRILLIN | | Authors: | Downing, A.K, Campbell, I.D, Handford, P.A. | | Deposit date: | 1996-08-05 | | Release date: | 1996-12-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Cell(Cambridge,Mass.), 85, 1996
|
|
2M9K
 
 | | RBPMS2-Nter | | Descriptor: | RNA-binding protein with multiple splicing 2 | | Authors: | Yang, Y. | | Deposit date: | 2013-06-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity.
Nucleic Acids Res., 42, 2014
|
|
3E94
 
 | |
2KCH
 
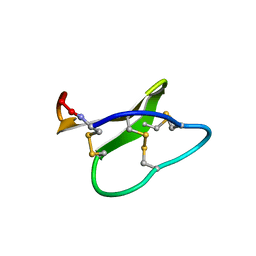 | | Solution structure of micelle-bound kalata B2 | | Descriptor: | Kalata-B2 | | Authors: | Wang, C.K. | | Deposit date: | 2008-12-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes.
Biophys.J., 97, 2009
|
|
1HZ4
 
 | | CRYSTAL STRUCTURE OF TRANSCRIPTION FACTOR MALT DOMAIN III | | Descriptor: | BENZOIC ACID, GLYCEROL, MALT REGULATORY PROTEIN, ... | | Authors: | Steegborn, C, Danot, O, Clausen, T, Huber, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of transcription factor MalT domain III: a novel helix repeat fold implicated in regulated oligomerization.
Structure, 9, 2001
|
|
2FJC
 
 | | Crystal structure of antigen TpF1 from Treponema pallidum | | Descriptor: | Antigen TpF1, FE (III) ION | | Authors: | Thumiger, A, Polenghi, A, Papinutto, E, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of antigen TpF1 from Treponema pallidum.
Proteins, 62, 2006
|
|
2L71
 
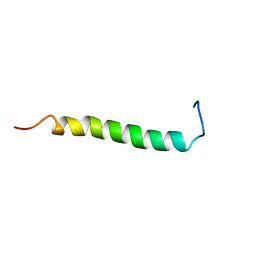 | | NMR solution structure of GIP in Bicellular media | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Venneti, K.C, Alana, I, O'Harte, F.P.M, Malthouse, P.J.G, Hewage, C.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational, receptor interaction and alanine scan studies of glucose-dependent insulinotropic polypeptide
Biochim.Biophys.Acta, 1814, 2011
|
|
3E4H
 
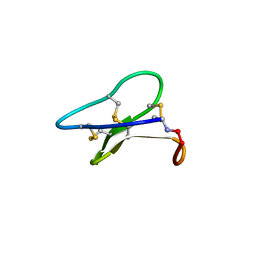 | | Crystal structure of the cyclotide varv F | | Descriptor: | Varv peptide F,Varv peptide F | | Authors: | Hu, S.H. | | Deposit date: | 2008-08-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined X-ray and NMR Analysis of the Stability of the Cyclotide Cystine Knot Fold That Underpins Its Insecticidal Activity and Potential Use as a Drug Scaffold
J.Biol.Chem., 284, 2009
|
|
2L70
 
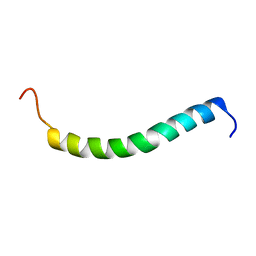 | | NMR solution structure of GIP in micellular media | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Venneti, K.C, Alana, I, O'Harte, F.P.M, Malthouse, P.J.G, Hewage, C.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational, receptor interaction and alanine scan studies of glucose-dependent insulinotropic polypeptide
Biochim.Biophys.Acta, 1814, 2011
|
|
2K7G
 
 | | Solution Structure of varv F | | Descriptor: | Varv peptide F | | Authors: | Wang, C.K. | | Deposit date: | 2008-08-10 | | Release date: | 2009-02-10 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold
J.Biol.Chem., 284, 2009
|
|
