1DBU
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
2LY7
 
 | | B-flap domain of RNA polymerase (B. subtilis) | | Descriptor: | DNA-directed RNA polymerase subunit beta | | Authors: | Mobli, M. | | Deposit date: | 2012-09-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase-induced remodelling of NusA produces a pause enhancement complex.
Nucleic Acids Res., 43, 2015
|
|
3KEW
 
 | | Crystal structure of probable alanyl-trna-synthase from Clostridium perfringens | | Descriptor: | DHHA1 domain protein, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alanyl-trna-synthase from Clostridium perfringens
To be Published
|
|
4UNK
 
 | | Crystal structure of human triosephosphate isomerase (mutant N15D) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Triosephosphate Isomerase (Mutant N15D)
To be Published
|
|
4UNL
 
 | | Crystal structure of a single mutant (N71D) of triosephosphate isomerase from human | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Single Mutant (N71D) of Triosephosphate Isomerase from Human
To be Published
|
|
3OQV
 
 | | AlbC, a cyclodipeptide synthase from Streptomyces noursei | | Descriptor: | AlbC, DITHIANE DIOL, PHOSPHATE ION | | Authors: | Sauguet, L, Ledu, M.H, Charbonnier, J.B, Gondry, M. | | Deposit date: | 2010-09-04 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis.
Nucleic Acids Res., 39, 2011
|
|
8BTZ
 
 | |
1NE9
 
 | | Crystal Structure of Weissella viridescens FemX at 1.7 Ang Resolution | | Descriptor: | FemX, MAGNESIUM ION | | Authors: | Biarrotte-Sorin, S, Maillard, A.P, Delettre, J, Sougakoff, W, Arthur, M, Mayer, C. | | Deposit date: | 2002-12-11 | | Release date: | 2004-02-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition.
Structure, 12, 2004
|
|
8BU8
 
 | |
6BMD
 
 | |
3OQJ
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQI
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQH
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC | | Descriptor: | GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OD1
 
 | | The crystal structure of an ATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C | | Descriptor: | ATP phosphoribosyltransferase regulatory subunit, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Bigelow, L, Hamilton, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of aATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C
To be Published
|
|
1WY5
 
 | | Crystal structure of isoluecyl-tRNA lysidine synthetase | | Descriptor: | Hypothetical UPF0072 protein AQ_1887 | | Authors: | Nakanishi, K, Fukai, S, Ikeuchi, Y, Soma, A, Sekine, Y, Suzuki, T, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Q11
 
 | | Crystal structure of an active fragment of human tyrosyl-tRNA synthetase with tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Yang, X.-L, Schimmel, P, Ribas de Pouplana, L. | | Deposit date: | 2003-07-18 | | Release date: | 2004-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3GS3
 
 | |
2NCI
 
 | |
4UV3
 
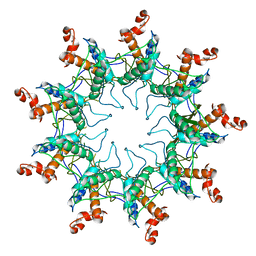 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4UV2
 
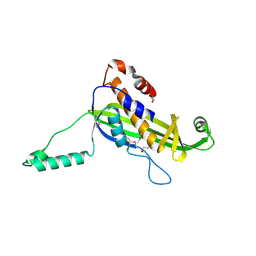 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
1QVA
 
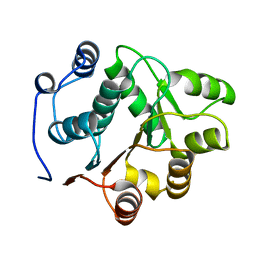 | | YEAST INITIATION FACTOR 4A N-TERMINAL DOMAIN | | Descriptor: | INITIATION FACTOR 4A | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase
RNA, 5, 1999
|
|
4H2W
 
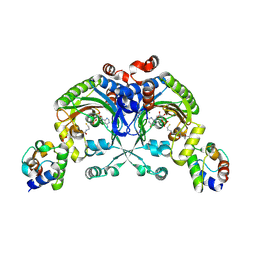 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4H2T
 
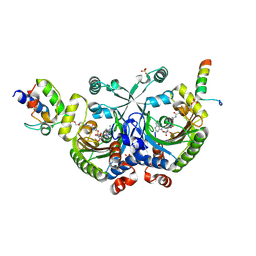 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and an analogue of glycyl adenylate | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-O-(glycylsulfamoyl)adenosine, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4C12
 
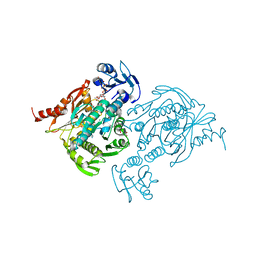 | | X-ray Crystal Structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fulop, V, Roper, D.I, Ruane, K.M, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity Determinants for Lysine Incorporation in Staphylococcus Aureus Peptidoglycan as Revealed by the Structure of a Mure Enzyme Ternary Complex.
J.Biol.Chem., 288, 2013
|
|
6EC7
 
 | |
