8HWK
 
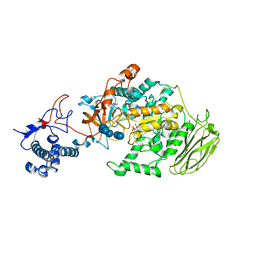 | | Limosilactobacillus reuteri N1 GtfB-maltohexaose | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
5WMV
 
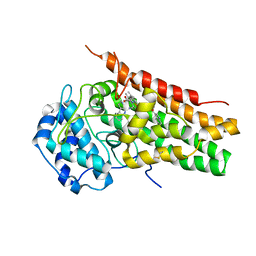 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WAU
 
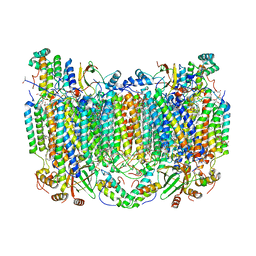 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Synchrotron X-Ray Crystallography at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Fromme, R, Ishigami, I, Yeh, S.Y, Zatsepin, N, Grant, T, Fromme, P, Rousseau, D. | | Deposit date: | 2017-06-27 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8HW3
 
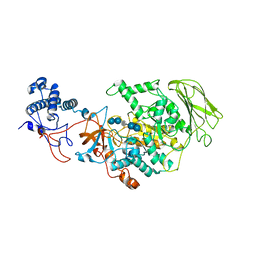 | | Limosilactobacillus reuteri N1 GtfB-acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8IHO
 
 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
8HYF
 
 | | Crystal Structure of Banana Lectin In-complex with Fucose at 2.95 A Resolution | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rasheed, S, Arif, R, Huda, N, Ahmad, M.S, Mateen, S.M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Banana Lectin in-Complex with L-Fucose at 2.95 A Resolution
To Be Published
|
|
6AYD
 
 | |
6B5A
 
 | |
6B4D
 
 | | Crystal structure of human carbonic anhydrase II in complex with a heteroaryl-pyrazole carboxylic acid derivative. | | Descriptor: | 3-(1-ethyl-1H-indol-3-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2017-09-26 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.196 Å) | | Cite: | Exploring Heteroaryl-pyrazole Carboxylic Acids as Human Carbonic Anhydrase XII Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6B1K
 
 | | Macrophage Migration Inhibitory Factor in Complex with a Naphthyridinone Inhibitor (3a) | | Descriptor: | 2-[1-(4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]-7-methyl-1,7-naphthyridin-8(7H)-one, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Krimmer, S.G, Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Adding a Hydrogen Bond May Not Help: Naphthyridinone vs Quinoline Inhibitors of Macrophage Migration Inhibitory Factor.
ACS Med Chem Lett, 8, 2017
|
|
6BDQ
 
 | |
6B59
 
 | |
7B9L
 
 | | MEK1 in complex with compound 23 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7BEA
 
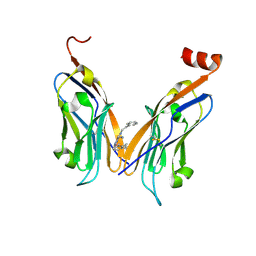 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | 2-(aminomethyl)-6-[(2-methyl-3-phenyl-phenyl)methoxy]-~{N}-(2-phenylethyl)imidazo[1,2-a]pyridin-3-amine, Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Butera, R, Wazynska, M, Holak, T, Domling, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Imidazopyridines as PD-1/PD-L1 Antagonists.
Acs Med.Chem.Lett., 12, 2021
|
|
6CJV
 
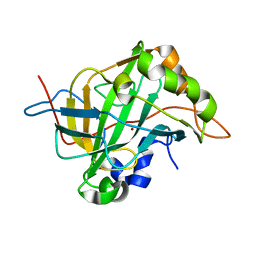 | | Carbonic anhydrase IX-mimic in complex with sucralose | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Carbonic anhydrase 2, ZINC ION | | Authors: | Lomelino, C.L, Murray, A.B, McKenna, R. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Sweet Binders: Carbonic Anhydrase IX in Complex with Sucralose.
ACS Med Chem Lett, 9, 2018
|
|
6CUP
 
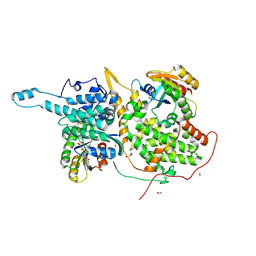 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | FORMIC ACID, GLYCEROL, GTPase HRas, ... | | Authors: | Phan, J, Abbott, J, Fesik, S.W. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Discovery of Quinazolines That Activate SOS1-Mediated Nucleotide Exchange on RAS.
ACS Med Chem Lett, 9, 2018
|
|
6CVX
 
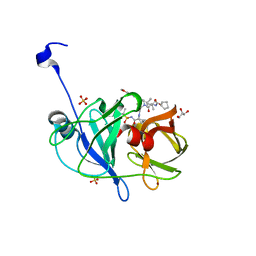 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-50 (MK-5172 linear analogue) | | Descriptor: | GLYCEROL, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-{[7-methoxy-3-(propan-2-yl)quinoxalin-2-yl]oxy}-L-prolinamide, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CYC
 
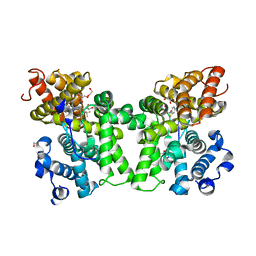 | | PDE2 in complex with compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 3-(hydroxymethyl)-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
6CVY
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-21 (MK-5172 linear analogue) | | Descriptor: | 3-methyl-N-[(pentyloxy)carbonyl]-L-valyl-(4R)-4-[(3-chloro-7-methoxyquinoxalin-2-yl)oxy]-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-L-prolinamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6BTP
 
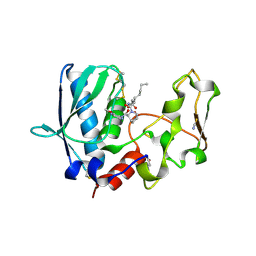 | | BMP1 complexed with a hydroxamate | | Descriptor: | (1R,3S,4S)-2-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]heptanoyl}-N-(3-methoxypyrazin-2-yl)-2-azabicyclo[2.2.1]heptane-3-carboxamide, Bone morphogenetic protein 1, THIOCYANATE ION, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
7B3W
 
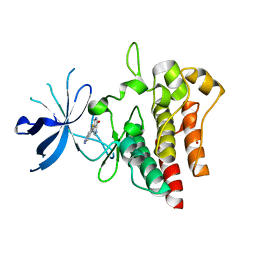 | | Crystal structure of c-MET bound by compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-fluoranyl-1~{H}-indazol-4-yl)-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
6CVW
 
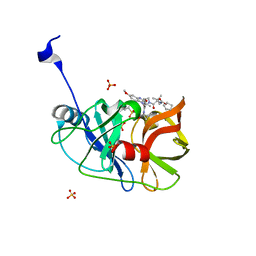 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-52 (MK-5172 linear analogue) | | Descriptor: | N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-L-prolinamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CDT
 
 | |
6CYB
 
 | | PDE2 in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,2,2-trifluoroethyl)-1-{(1S)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design and Procognitive Assessment of a Potent and Selective Phosphodiesterase 2A Inhibitor.
ACS Med Chem Lett, 9, 2018
|
|
7C2V
 
 | | Crystal Structure of IRAK4 kinase in complex with the inhibitor CA-4948 | | Descriptor: | 2-(2-methylpyridin-4-yl)-N-[2-morpholin-4-yl-5-[(3R)-3-oxidanylpyrrolidin-1-yl]-[1,3]oxazolo[4,5-b]pyridin-6-yl]-1,3-oxazole-4-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Krishnamurthy, N.R, Robert, B. | | Deposit date: | 2020-05-09 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of CA-4948, an Orally Bioavailable IRAK4 Inhibitor for Treatment of Hematologic Malignancies.
Acs Med.Chem.Lett., 11, 2020
|
|
