6GAU
 
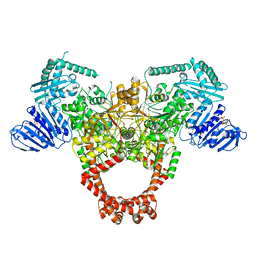 | | Extremely 'open' clamp structure of DNA gyrase: role of the Corynebacteriales GyrB specific insert | | Descriptor: | DNA gyrase subunit B,DNA gyrase subunit A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Petrella, S, Capton, E, Alzari, P.M, Aubry, A, Mayer, C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity.
Structure, 27, 2019
|
|
9IMJ
 
 | |
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
2IOP
 
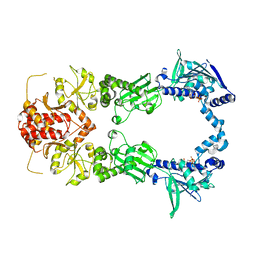 | | Crystal Structure of Full-length HtpG, the Escherichia coli Hsp90, Bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG | | Authors: | Shiau, A.K, Harris, S.F, Agard, D.A. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements.
Cell(Cambridge,Mass.), 127, 2006
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
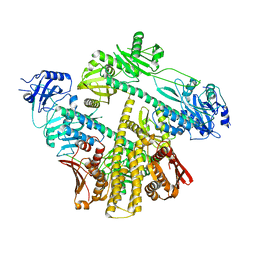 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
9EPN
 
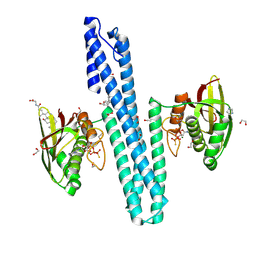 | | Crystal structure of HprS histidine kinase cytoplasmic fragment from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, GLYCOCHOLIC ACID, IMIDAZOLE, ... | | Authors: | Koczurowska, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2024-03-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biophysical characterization of the cytoplasmic domains of HprS kinase and its interactions with the cognate regulator HprR.
Arch.Biochem.Biophys., 764, 2024
|
|
9JFJ
 
 | | Crystal structure of the cytoplasmic domain of ZraS in ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sensor histidine kinase ZraS | | Authors: | Mahapatra, N, Pandey, S, Mahanta, P, Acharya, R. | | Deposit date: | 2024-09-04 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Insights Into the Conformational Dynamics of the Cytoplasmic Domain of Metal-Sensing Sensor Histidine Kinase ZraS.
Proteins, 2025
|
|
3DGE
 
 | |
2PNR
 
 | | Crystal Structure of the asymmetric Pdk3-l2 Complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3 | | Authors: | Vassylyev, D.G, Steussy, C.N, Devedjiev, Y. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Asymmetric complex of Pyruvate Dehydrogenase
Kinase 3 with Lipoyl domain 2 and its Biological Implications
J.Mol.Biol., 370, 2007
|
|
6ZT5
 
 | | Complex between a homodimer of Mycobacterium smegmatis MfpA and a single copy of the N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit | | Descriptor: | DNA gyrase subunit B, Pentapeptide repeat protein MfpA, SULFATE ION | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZT3
 
 | | N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit complexed with ADPNP | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZY8
 
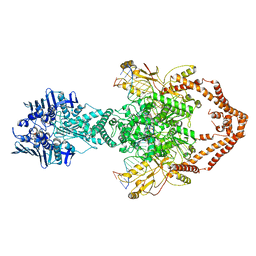 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY7
 
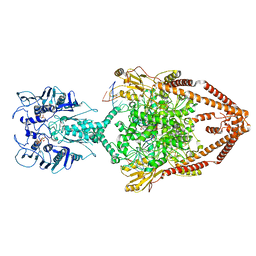 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY6
 
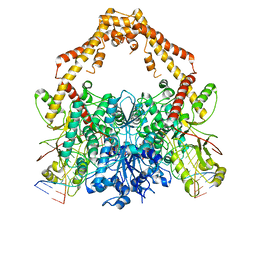 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY5
 
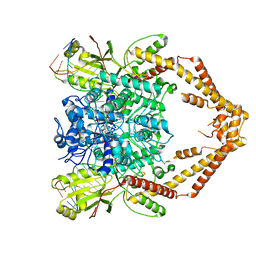 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
2CG9
 
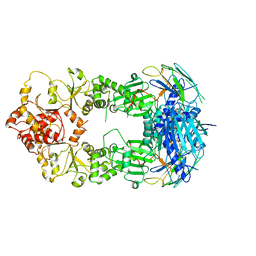 | | Crystal structure of an Hsp90-Sba1 closed chaperone complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CO-CHAPERONE PROTEIN SBA1 | | Authors: | Ali, M.M.U, Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of an Hsp90-Nucleotide-P23/Sba1 Closed Chaperone Complex
Nature, 440, 2006
|
|
2Q8I
 
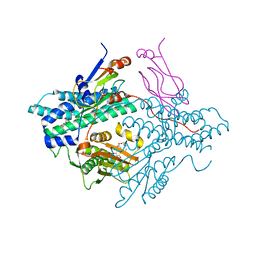 | | Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, GLYCEROL, ... | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2CH4
 
 | |
1Y8P
 
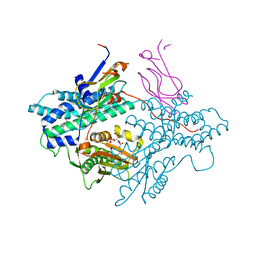 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8N
 
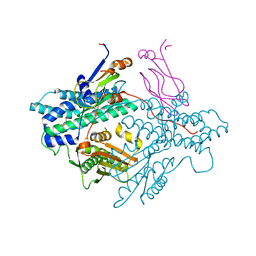 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, POTASSIUM ION, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8O
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y4U
 
 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1Y4S
 
 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
