1KJF
 
 | |
1KJG
 
 | |
1KJH
 
 | |
1KNA
 
 | |
1KNE
 
 | |
1KNO
 
 | |
1KNV
 
 | | Bse634I restriction endonuclease | | Descriptor: | ACETATE ION, Bse634I restriction endonuclease, CHLORIDE ION | | Authors: | Grazulis, S, Deibert, M, Rimseliene, R, Skirgaila, R, Sasnauskas, G, Lagunavicius, A, Repin, V, Urbanke, C, Huber, R, Siksnys, V. | | Deposit date: | 2001-12-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence.
Nucleic Acids Res., 30, 2002
|
|
1KO2
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with an oxidized Cys (cysteinesulfonic) | | Descriptor: | ACETATE ION, VIM-2 metallo-beta-lactamase, ZINC ION | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1KO3
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with Cys221 reduced | | Descriptor: | ACETATE ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KR7
 
 | | Crystal structure of the nerve tissue mini-hemoglobin from the nemertean worm Cerebratulus lacteus | | Descriptor: | ACETATE ION, Neural globin, OXYGEN MOLECULE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Geuens, E, Yamauchi, k, Ascenzi, P, Riggs, A.F, Moens, L, Bolognesi, M. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 109 residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the alpha-helical globin fold
Structure, 10, 2002
|
|
1KU5
 
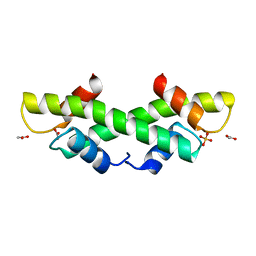 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
1KWG
 
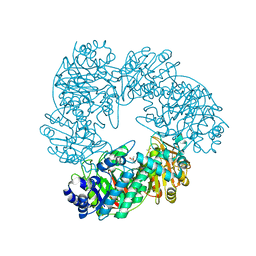 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
1KWK
 
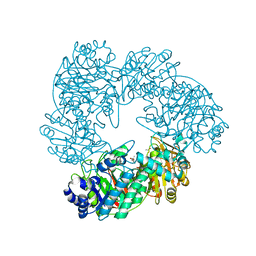 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
1KX3
 
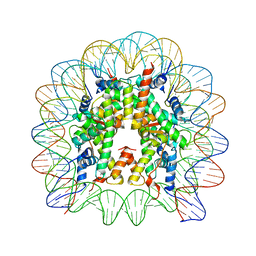 | | X-Ray Structure of the Nucleosome Core Particle, NCP146, at 2.0 A Resolution | | Descriptor: | DNA (5'(ATCAATATCCACCTGCAGATTCTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGAATTCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGAATCTGCAGGTGGATATTGAT)3'), MANGANESE (II) ION, histone H2A.1, ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
1KX4
 
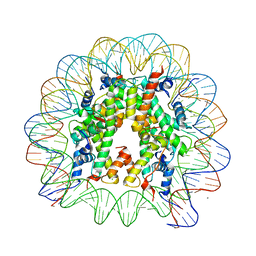 | | X-Ray Structure of the Nucleosome Core Particle, NCP146b, at 2.6 A Resolution | | Descriptor: | CHLORIDE ION, DNA (5'(ATCTCCAAATATCCCTTGCGGATCGTAGAAAAAGTGTGTCAAACTGCGCTATCAAAGGGAAACTTCAACTGAATTCAGTTGAAGTTTCCCTTTGATAGCGCAGTTTGACACACTTTTTCTACGATCCGCAAGGGATATTTGGAGAT)3'), MANGANESE (II) ION, ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
1KX5
 
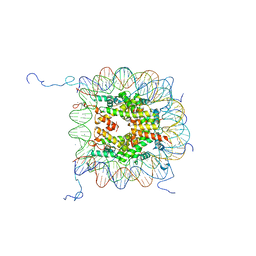 | | X-Ray Structure of the Nucleosome Core Particle, NCP147, at 1.9 A Resolution | | Descriptor: | CHLORIDE ION, DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGAATCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGATTCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
1KX9
 
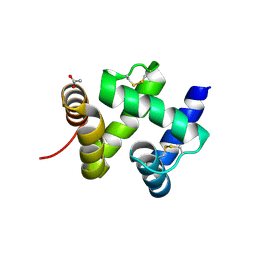 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1L0V
 
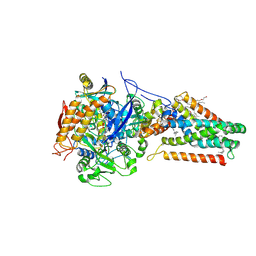 | | Quinol-Fumarate Reductase with Menaquinol Molecules | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1L5J
 
 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
1L6R
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1L8S
 
 | |
1L9A
 
 | | CRYSTAL STRUCTURE OF SRP19 IN COMPLEX WITH THE S DOMAIN OF SIGNAL RECOGNITION PARTICLE RNA | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN, ... | | Authors: | Oubridge, C, Kuglstatter, A, Jovine, L, Nagai, K. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle.
Mol.Cell, 9, 2002
|
|
1LDG
 
 | | PLASMODIUM FALCIPARUM L-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C, Banfield, M, Barker, J, Higham, C, Moreton, K, Turgut-Balik, D, Brady, L, Holbrook, J.J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design.
Nat.Struct.Biol., 3, 1996
|
|
1LDM
 
 | |
