5W7B
 
 | | Rabbit acyloxyacyl hydrolase (AOAH), proteolytically processed, S262A mutant, with LPS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7W4Q
 
 | | Deactive state CI from Q1-NADH dataset, Subclass 6 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4F
 
 | | Active state CI from Q1-NADH dataset, Subclass 4 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-27 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W62
 
 | | Mevo lectin complex with mannotetraose (Man4) | | Descriptor: | 1,2-ETHANEDIOL, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Vijayan, M. | | Deposit date: | 2021-12-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mevo lectin complex with mannotetraose (Man4)
To Be Published
|
|
7WQV
 
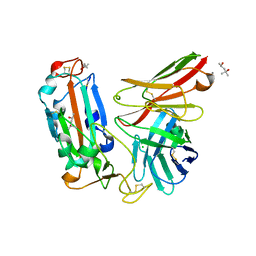 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTI
 
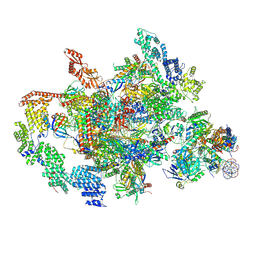 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58hex) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XT7
 
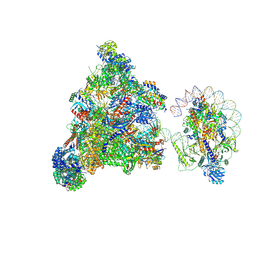 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49B) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
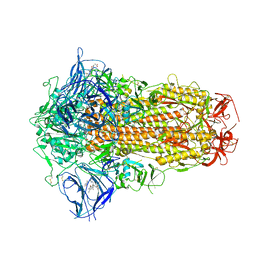 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
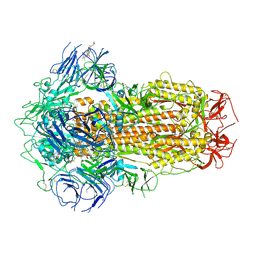 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
5WK5
 
 | |
7YAT
 
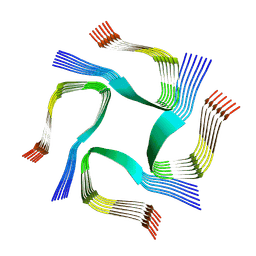 | | CryoEM tetra protofilament structure of the hamster prion 108-144 fibril | | Descriptor: | Major prion protein | | Authors: | Chen, E.H.-L, Kao, S.-W, Lee, C.-H, Huang, J.Y.C, Chen, R.P.-Y, Wu, K.-P. | | Deposit date: | 2022-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 angstrom Cryo-EM Tetra-Protofilament Structure of the Hamster Prion 108-144 Fibril Reveals an Ordered Water Channel in the Center.
J.Am.Chem.Soc., 144, 2022
|
|
5W75
 
 | |
5W9H
 
 | |
5WBN
 
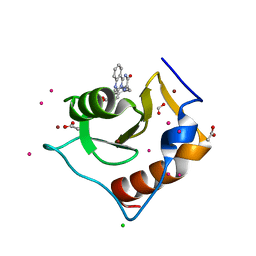 | | Crystal structure of fragment 3-(3-Benzyl-2-oxo-2H-[1,2,4]triazino[2,3-c]quinazolin-6-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-benzyl-2-oxo-2H-[1,2,4]triazino[2,3-c]quinazolin-6-yl)propanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Harding, R.J, Walker, J.R, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of fragment 3-(3-Benzyl-2-oxo-2H-[1,2,4]triazino[2,3-c]quinazolin-6-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain
To be published
|
|
7Y6S
 
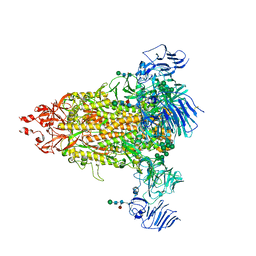 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
5WHU
 
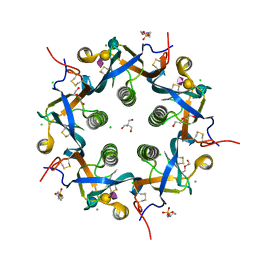 | | Crystal structure of 3'SL bound ArtB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ArtB protein, ... | | Authors: | Gao, X, Galan, J.E. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
5WJW
 
 | | Cryo-EM structure of B. subtilis flagellar filaments H84R | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJX
 
 | | Cryo-EM structure of B. subtilis flagellar filaments S17P | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WNT
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | (1R,2S,3S,4R,6R)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-alpha-L-altropyranosyl)-beta-L-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-allopyranoside, 16S Ribosomal RNA rRNA, MAGNESIUM ION, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WNV
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | (1R,2S,3S,4R,6R)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-alpha-L-altropyranosyl)-beta-L-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-allopyranoside, 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7XST
 
 | |
