8C5H
 
 | |
8PUU
 
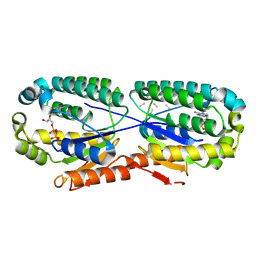 | | Giardia intestinalis deoxyadenosine kinase forms a functional tetramer | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Deoxynucleoside kinase | | Authors: | Ranjbarian, F, Rafie, K, Shankar, K, Krakovka, S, Svaerd, S, Carlson, L.-A, Hofer, A. | | Deposit date: | 2023-07-17 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization of deoxyadenosine kinase meets the demands of a DNA replication substrate challenge in Giardia intestinalis.
Nucleic Acids Res., 52, 2024
|
|
8BPD
 
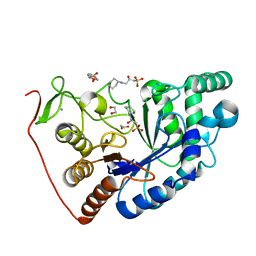 | | Structural and Functional Characterization of the Novel Endo-alpha(1,4)-Fucoidanase Mef1 from the Marine Bacterium Muricauda eckloniae | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Mikkelsen, M.D, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of the novel endo-alpha (1,4)-fucoidanase Mef1 from the marine bacterium Muricauda eckloniae.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BQ7
 
 | |
7APR
 
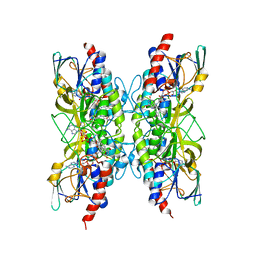 | | Bacillithiol Disulfide Reductase Bdr (YpdA) from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YpdA family putative bacillithiol disulfide reductase Bdr | | Authors: | Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase.
Biochemistry, 59, 2020
|
|
8BWK
 
 | |
8R43
 
 | |
8BZK
 
 | |
8C0M
 
 | |
7B6B
 
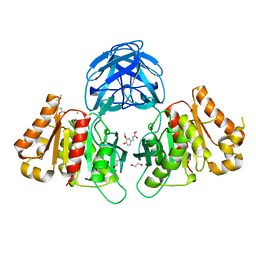 | | The carbohydrate binding module family 48 (CBM48) and carboxy-terminal carbohydrate esterase family 1 (CE1) domains of the multidomain esterase DmCE1B from Dysgonomonas mossii in complex with methyl ferulate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48, ... | | Authors: | Mazurkewich, S, Kmezik, C, Branden, G, Larsbrink, J. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A polysaccharide utilization locus from the gut bacterium Dysgonomonas mossii encodes functionally distinct carbohydrate esterases.
J.Biol.Chem., 296, 2021
|
|
8C67
 
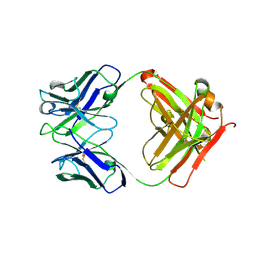 | | Crystal structure of Ab25 Fab | | Descriptor: | antibody 25 heavy chain, antibody 25 light chain | | Authors: | Nyblom, M, Izadi, A, Tang, D, Bahnan, W, Happonen, L, Malmstroem, J, Shannon, O, Malmstroem, L, Nordenfelt, P. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The hinge-engineered IgG1-IgG3 hybrid subclass IgGh 47 potently enhances Fc-mediated function of anti-streptococcal and SARS-CoV-2 antibodies.
Nat Commun, 15, 2024
|
|
8BVX
 
 | |
8R4U
 
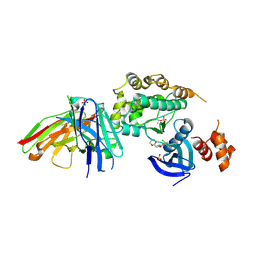 | | Structure of salt-inducible kinase 3 with inhibitors | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
8R4Q
 
 | | Salt inducible kinase 3 in complex with inhibitor | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.838 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
7B5V
 
 | | The carbohydrate binding module family 48 (CBM48) and carboxy-terminal carbohydrate esterase family 1 (CE1) domains of the multidomain esterase DmCE1B from Dysgonomonas mossii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48, ... | | Authors: | Mazurkewich, S, Kmezik, C, Branden, G, Larsbrink, J. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A polysaccharide utilization locus from the gut bacterium Dysgonomonas mossii encodes functionally distinct carbohydrate esterases.
J.Biol.Chem., 296, 2021
|
|
8CB5
 
 | |
8CB8
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with tetrapeptide Ser-Thr-Leu-Leu | | Descriptor: | Listeriolysin regulatory protein, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Oelker, M, Lindgren, C, Sauer-Eriksson, A.E. | | Deposit date: | 2023-01-25 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of promiscuous inhibition of Listeria virulence activator PrfA by oligopeptides.
Cell Rep, 44, 2025
|
|
7B65
 
 | | Structure of NUDT15 R139C Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B64
 
 | | Structure of NUDT15 V18I Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B63
 
 | | Structure of NUDT15 in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B66
 
 | | Structure of NUDT15 R139H Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
8CBP
 
 | |
7AVG
 
 | | Perdeuterated hen egg-white lysozyme at 100 K | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
7AOP
 
 | | Structure of NUDT15 in complex with inhibitor TH8321 | | Descriptor: | 2-azanyl-9-cyclohexyl-8-(2-methoxyphenyl)-3~{H}-purine-6-thione, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
8Q8H
 
 | |
