7REP
 
 | |
3DBM
 
 | | Crystal Structure of Allene oxide synthase | | Descriptor: | (9E,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-06-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5ZI0
 
 | | Ketoreductase LbCR mutant - M8 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Gong, X.M, Zheng, G.W, Xu, J.H. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Development of an Engineered Ketoreductase with Simultaneously Improved Thermostability and Activity for Making a Bulky Atorvastatin Precursor
Acs Catalysis, 9, 2019
|
|
6K9D
 
 | | glycoside hydrolase family 12 (GH12) englucanase | | Descriptor: | GH12 beta-1, 4-endoglucanase | | Authors: | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, Yingguo, B, Bin, Y. | | Deposit date: | 2019-06-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
3IJ9
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
5ZJP
 
 | |
4PSA
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-N1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
3IJQ
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4PSK
 
 | | Mycobacterium tuberculosis RecA phosphate bound low temperature structure I-LT | | Descriptor: | PHOSPHATE ION, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
5Z3B
 
 | | Glycosidase Y48F | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
7REO
 
 | |
6K35
 
 | | Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-N-acetylglucosaminidase Nag2 | | Authors: | Meekrathok, P, Stubbs, K.A, Bulmer, D.M, van den Berg, B, Suginta, W. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Febs J., 287, 2020
|
|
5ZLH
 
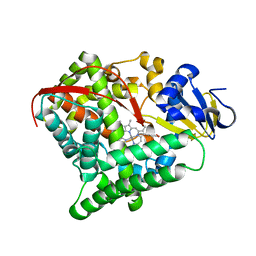 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
2UUQ
 
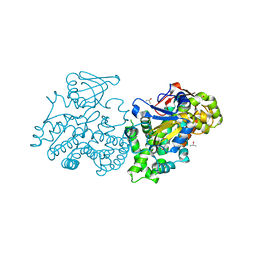 | |
2VE4
 
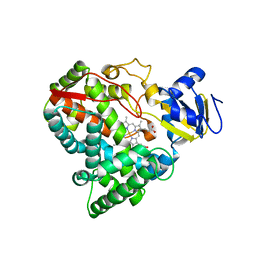 | | Substrate free cyanobacterial CYP120A1 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 120 | | Authors: | Kuhnel, K, Ke, N, Sligar, S.G, Schuler, M.A, Schlichting, I. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Substrate-Free and Retinoic Acid-Bound Cyanobacterial Cytochrome P450 Cyp120A1.
Biochemistry, 47, 2008
|
|
4B49
 
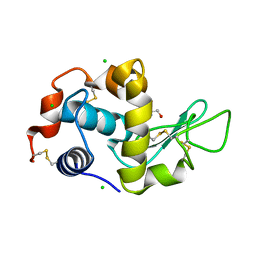 | | 1.15 A Structure of Lysozyme Crystallized without 2-methyl-2,4- pentanediol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, LYSOZYME C | | Authors: | Jakoncic, J, Berger, J, Stauber, M, Axelbaum, A, Asherie, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallization of Lysozyme with (R)-, (S)- and (Rs)-2-Methyl-2,4-Pentanediol
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3IO1
 
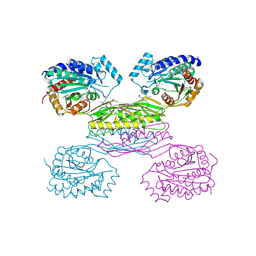 | | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae | | Descriptor: | Aminobenzoyl-glutamate utilization protein, SODIUM ION, YTTRIUM (III) ION | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-13 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae
To be Published
|
|
2GU0
 
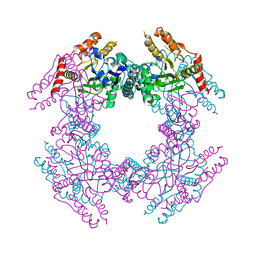 | |
2GO1
 
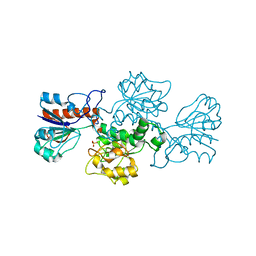 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 | | Descriptor: | NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Stekhanova, T.N, Boiko, K.M, Popov, V.O. | | Deposit date: | 2006-04-12 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 A
Crystallography reports, 50, 2005
|
|
3DHG
 
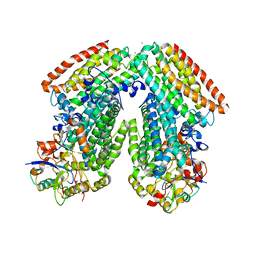 | | Crystal Structure of Toluene 4-Monoxygenase Hydroxylase | | Descriptor: | AZIDE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D9F
 
 | | Nitroalkane oxidase: active site mutant S276A crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
4K5Y
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, ... | | Authors: | Hollenstein, K, Kean, J, Bortolato, A, Cheng, R.K.Y, Dore, A.S, Jazayeri, A, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2013-04-15 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.977 Å) | | Cite: | Structure of class B GPCR corticotropin-releasing factor receptor 1.
Nature, 499, 2013
|
|
5Z7M
 
 | | SmChiA sliding-intermediate with chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase A, ... | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
3DK8
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
