4Y7T
 
 | | Structural analysis of MurU | | Descriptor: | GLYCEROL, Nucleotidyl transferase, SULFATE ION | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7U
 
 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, GLYCEROL, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7V
 
 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, IMIDODIPHOSPHORIC ACID, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4YDW
 
 | |
5A0O
 
 | | adhiron raised against p300 | | Descriptor: | ADHIRON | | Authors: | Kyle, H.F, Wickson, K.F, Stott, J, Burslem, G.M, Breeze, A.L, Tiede, C, Tomlinson, D.C, Warriner, S.L, Nelson, A, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Exploration of the Hif-1Alpha.P300 Interface Using Peptide and Adhiron Phage Display Technologies
Mol.Biosyst., 11, 2015
|
|
5AEI
 
 | | Designed Armadillo repeat protein YIIIM5AII in complex with peptide (KR)5 | | Descriptor: | ACETATE ION, CALCIUM ION, DESIGNED ARMADILLO REPEAT PROTEIN YIIIM5AII, ... | | Authors: | Hansen, S, Tremmel, D, Madhurantakam, C, Reichen, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2015-08-31 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Energetic Contributions of a Designed Modular Peptide-Binding Protein with Picomolar Affinity.
J.Am.Chem.Soc., 138, 2016
|
|
3K1W
 
 | | New Classes of Potent and Bioavailable Human Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetylamino-2-deoxy-alpha-L-idopyranose, 4-{4-[3-(2-bromo-5-fluorophenoxy)propyl]phenyl}-N-(2-chlorobenzyl)-N-cyclopropyl-1,2,5,6-tetrahydropyridine-3-carboxamide, ... | | Authors: | Prade, L. | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New classes of potent and bioavailable human renin inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2DX3
 
 | |
5LE4
 
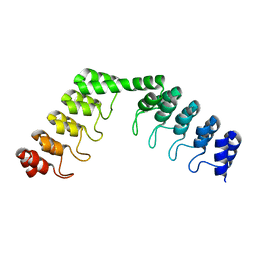 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_11_D12 | | Descriptor: | DD_D12_11_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEA
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_12_3G124 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DD_Off7_12_3G124 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LED
 
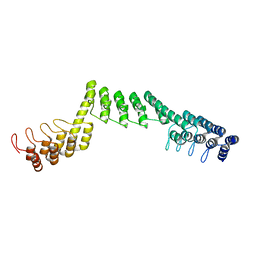 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
3JCA
 
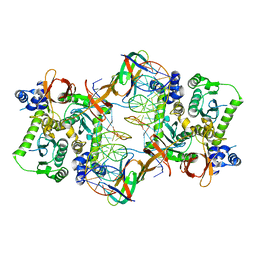 | | Core model of the Mouse Mammary Tumor Virus intasome | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*CP*GP*CP*AP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*CP*TP*G)-3', 5'-D(*CP*AP*GP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*TP*GP*CP*GP*GP*CP*A)-3', Integrase, ... | | Authors: | Lyumkis, D.L, Ballandras-Colas, A, Brown, M, Cook, N.J, Dewdney, T.G, Demeler, B, Cherepanov, P, Engelman, A.N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
5MFJ
 
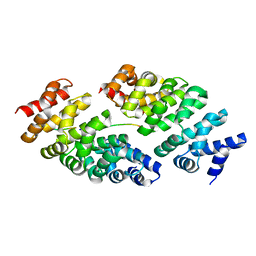 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)5 | | Descriptor: | (KR)5, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
3JAF
 
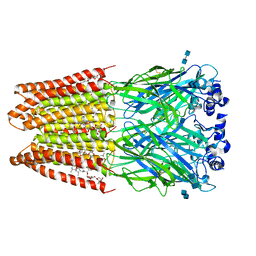 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.801 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
5MFC
 
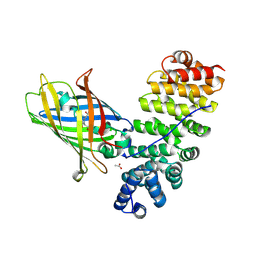 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5MFL
 
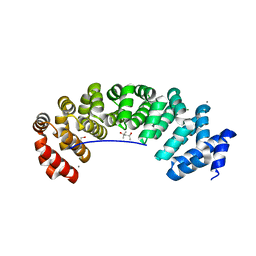 | | Designed armadillo repeat protein (KR)5_GS10_YIIIM6AII | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (KR)5_GS10_YIIIM6AII, 1,2-ETHANEDIOL, ... | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFK
 
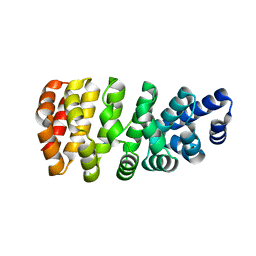 | | Designed armadillo repeat protein YIII(Dq.V1)4CPAF in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V1)4CPAF | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
2E4E
 
 | |
2R2V
 
 | |
5MFF
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, 1,2-ETHANEDIOL, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFN
 
 | | Designed armadillo repeat protein YIIIM5AII | | Descriptor: | CALCIUM ION, D-MALATE, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
2DX2
 
 | |
5MFB
 
 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFH
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)5 | | Descriptor: | (RR)5, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
