3EY4
 
 | | Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11-hydroxysteroid dehydrogenase type 1 (11-HSD1) inhibitors: Reducing pregnane X receptor (PXR) activity and exploring activity in a monkey pharmacodynamic model | | Descriptor: | (5S)-2-{[(1S)-1-(4-fluorophenyl)ethyl]amino}-5-(1-hydroxy-1-methylethyl)-5-methyl-1,3-thiazol-4(5H)-one, 11-beta-Hydroxysteroid Dehydrogenase 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J.D, Jordan, S.R, Li, V. | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11-hydroxysteroid dehydrogenase type 1 (11-HSD1) inhibitors: Reducing pregnane X receptor (PXR) activity and exploring activity in a monkey pharmacodynamic model
To be Published, 2008
|
|
4NDN
 
 | | Structural insights of MAT enzymes: MATa2b complexed with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
5KIJ
 
 | | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex | | Descriptor: | 1,4-BUTANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, ... | | Authors: | Karaveg, K, Xiang, Y, Moremen, K.W. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Substrate recognition and catalysis by GH47 alpha-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4S2O
 
 | | OXA-10 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, COBALT (II) ION | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
1X26
 
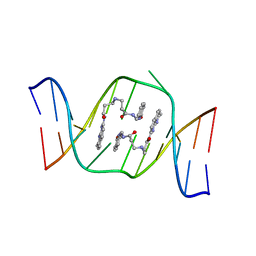 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | Descriptor: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | Authors: | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | Deposit date: | 2005-04-20 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
4RPB
 
 | |
5NIU
 
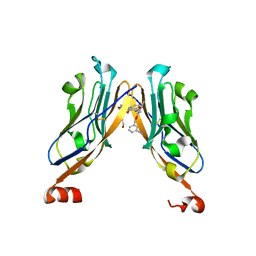 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{R})-2-[[2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-5-methyl-phenyl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells.
Oncotarget, 8, 2017
|
|
3D5Q
 
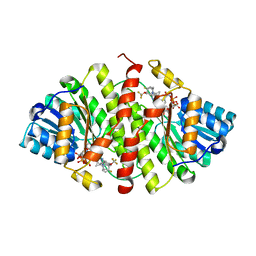 | | Crystal Structure of 11b-HSD1 in Complex with Triazole Inhibitor | | Descriptor: | 3-[1-(4-fluorophenyl)cyclopropyl]-4-(1-methylethyl)-5-[4-(trifluoromethoxy)phenyl]-4H-1,2,4-triazole, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P.C. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Distinctive molecular inhibition mechanisms for selective inhibitors of human 11beta-hydroxysteroid dehydrogenase type 1.
Bioorg.Med.Chem., 16, 2008
|
|
4S2I
 
 | | CTX-M-15 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
5HVB
 
 | | 5-methyl-6-(1-naphthylthio)thieno[2,3-d]pyrimidine 2,4-diamine | | Descriptor: | 5-methyl-6-(naphthalen-1-ylsulfanyl)thieno[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2016-01-28 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human dihydrofolate reductase ternary complex with a series of fluorine substituted 5-methyl-6-(1-napthylthio)[2,3-d]pyrimidine-2,4-diamines
To Be Published
|
|
5N4R
 
 | |
6CDP
 
 | | Vaccine-elicited HIV-1 neutralizing antibody vFP20.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP20.01 Fab heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
5HGJ
 
 | | Structure of integrin alpha1beta1 and alpha2beta1 I-domains explain differential calcium-mediated ligand recognition | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Brown, K.L, Banerjee, S, Feigley, A, Abe, H, Blackwell, T, Zent, R, Pozzi, A, Hudson, B.H. | | Deposit date: | 2016-01-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Salt-bridge modulates differential calcium-mediated ligand binding to integrin alpha 1- and alpha 2-I domains.
Sci Rep, 8, 2018
|
|
5KK7
 
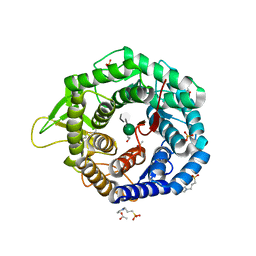 | | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex | | Descriptor: | 1,4-BUTANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, ... | | Authors: | Karaveg, K, Xiang, Y, Moremen, K.W. | | Deposit date: | 2016-06-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7324 Å) | | Cite: | Substrate recognition and catalysis by GH47 alpha-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5W6D
 
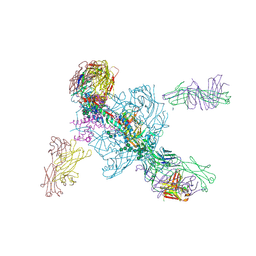 | | Crystal structure of BG505-SOSIP.v4.1-GT1-N137A in complex with Fabs 35022 and 9H/109L | | Descriptor: | 109L FAB light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garces, F, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo.
J. Exp. Med., 214, 2017
|
|
3TD8
 
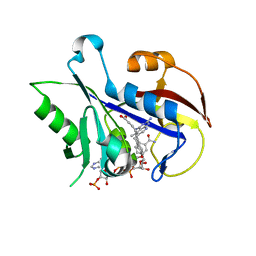 | | Structural Analysis of Pneumocystis carinii Dihydrofolate Reductase Complex with NADPH and 2,4-diamino-5-methyl-6-[2'-(4-carboxy-1-pentynyl)-5'-methoxybenzyl]pyrido[2,3-d]pyrimidine | | Descriptor: | 6-{2-[(2,4-diamino-5-methylpyrido[2,3-d]pyrimidin-6-yl)methyl]-4-methoxyphenyl}hex-5-ynoic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Stewart, E. | | Deposit date: | 2011-08-10 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Pneumocystis cariniidihydrofolate reductase complexed with NADPH and 2,4-diamino-6-[2-(5-carboxypent-1-yn-1-yl)-5-methoxybenzyl]-5-methylpyrido[2,3-d]pyrimidine.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5WDU
 
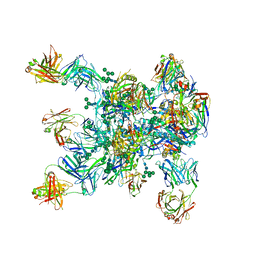 | | HIV-1 Env BG505 SOSIP.664 H72C-H564C trimer in complex with bNAbs PGT122 Fab, 35O22 Fab and NIH45-46 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Julien, J.-P, Torrents de la Pena, A, Sanders, R.W, Wilson, I.A. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Improving the Immunogenicity of Native-like HIV-1 Envelope Trimers by Hyperstabilization.
Cell Rep, 20, 2017
|
|
5CIL
 
 | | Crystal Structure of non-neutralizing version of 4E10 (WDWD) with epitope bound | | Descriptor: | ACETATE ION, CHLORIDE ION, FAB 4E10 HEAVY CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
6DTN
 
 | |
6DAS
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | N-[(1R)-6-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)-2,3-dihydro-1H-inden-1-yl]-3-methoxy-4-methylbenzamide, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
1HFC
 
 | |
6EKX
 
 | |
6H14
 
 | | Crystal structure of TcACHE complexed to 1-(6-oxo-1,2,3,4,6,10b-hexahydropyrido[2,1-a]isoindol-10-yl)-3-(4-(1-(2-((1,2,3,4-tetrahydroacridin-9-yl)amino)ethyl)-1H-1,2,3-triazol-4-yl)pyridin-2-yl)urea | | Descriptor: | 1,2-ETHANEDIOL, 1-[(10~{b}~{S})-6-oxidanylidene-2,3,4,10~{b}-tetrahydro-1~{H}-pyrido[2,1-a]isoindol-10-yl]-3-[4-[1-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pyridin-2-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2018-07-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design, biological evaluation and X-ray crystallography of nanomolar multifunctional ligands targeting simultaneously acetylcholinesterase and glycogen synthase kinase-3.
Eur.J.Med.Chem., 168, 2019
|
|
4DUN
 
 | | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, Putative phenazine biosynthesis PhzC/PhzF protein, ... | | Authors: | Brunzelle, J.S, Wawrzak, W, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76A X-ray Crystal Structure of a Putative Phenazine Biosynthesis PhzC/PhzF Protein from Clostridium difficile (strain 630)
To be Published
|
|
5M17
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
