7HRM
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z445135574 | | Descriptor: | 1-benzothiophen-6-amine 1,1-dioxide, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QJW
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z198194396 | | Descriptor: | 1,2-ETHANEDIOL, 4-(furan-2-carbonyl)piperazine-1-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QQI
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-178-994 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-[3-(methoxymethyl)phenyl]-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3RME
 
 | | AMCase in complex with Compound 5 | | Descriptor: | Acidic mammalian chitinase, GLYCEROL, N-ethyl-2-(4-methylpiperazin-1-yl)pyridine-3-carboxamide | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
4L02
 
 | | Crystal Structure of SphK1 with inhibitor | | Descriptor: | (2R,4S)-1-[2-(4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)ethyl]-2-(hydroxymethyl)piperidin-4-ol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N, Wang, Z. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure guided design of a series of sphingosine kinase (SphK) inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5QQD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with Z56880342 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QT2
 
 | | PanDDA analysis group deposition -- Partial occupancy interpretation of PanDDA event map: SETDB1 in complex with FMOPL000074a | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(1~{H}-pyrazol-3-yl)phenoxy]pyrimidine, DIMETHYL SULFOXIDE, ... | | Authors: | Harding, R.J, Tempel, W, DOUANGAMATH, A, BRANDAO-NETO, J, Collins, P.M, Krojer, T, Mader, P, Schapira, M, von Delft, F, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Santhakumar, V. | | Deposit date: | 2019-06-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4BAF
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethyltriazoledipicolinate complex at 1.51 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
2OW6
 
 | | Golgi alpha-mannosidase II complex with (1r,5s,6s,7r,8s)-1-thioniabicyclo[4.3.0]nonan-5,7,8-triol chloride | | Descriptor: | (1R,5S,6S,7R,8S)-1-THIONIABICYCLO[4.3.0]NONAN-5,7,8-TRIOL, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A. | | Deposit date: | 2007-02-15 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Binding of sulfonium-ion analogues of di-epi-swainsonine and 8-epi-lentiginosine to Drosophila Golgi alpha-mannosidase II: The role of water in inhibitor binding.
Proteins, 71, 2008
|
|
5QTO
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with 1R-0641 | | Descriptor: | 1,2-ETHANEDIOL, 3-(difluoromethyl)-8-(trifluoromethyl)[1,2,4]triazolo[4,3-a]pyridine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTP
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with AE-0227 | | Descriptor: | 1,2-ETHANEDIOL, 8-chloro-6-(trifluoromethyl)imidazo[1,2-a]pyridine-7-carbonitrile, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2EQM
 
 | | Solution structure of the TUDOR domain of PHD finger protein 20-like 1 [Homo sapiens] | | Descriptor: | PHD finger protein 20-like 1 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TUDOR domain of PHD finger protein 20-like 1 [Homo sapiens]
To be Published
|
|
2B4E
 
 | |
1Q92
 
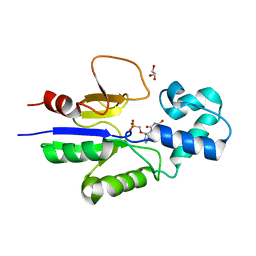 | | Crystal structure of human mitochondrial deoxyribonucleotidase in complex with the inhibitor PMcP-U | | Descriptor: | 5(3)-deoxyribonucleotidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rinaldo-Matthis, A, Rampazzo, C, Balzarini, J, Reichard, P, Bianchi, V, Nordlund, P. | | Deposit date: | 2003-08-22 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the mitochondrial deoxyribonucleotidase in complex with two specific inhibitors
Mol.Pharmacol., 65, 2004
|
|
5QU8
 
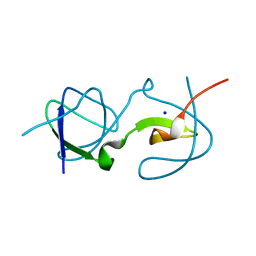 | |
4HA4
 
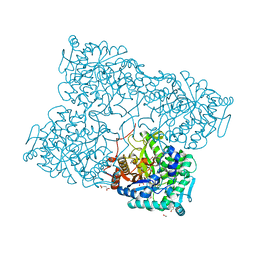 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with glycerol | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-galactosidase, GLYCEROL, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
4HAW
 
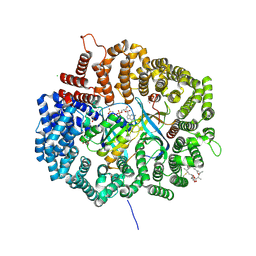 | |
4L71
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-A on the Ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.900001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3RXO
 
 | | Crystal structure of Trypsin complexed with (3-pyrrol-1-ylphenyl)methanamine | | Descriptor: | 1-[3-(1H-pyrrol-1-yl)phenyl]methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
4KV8
 
 | | Crystal structure of HIV RT in complex with BILR0355BS | | Descriptor: | 11-ethyl-5-methyl-8-[2-(1-oxidanylquinolin-4-yl)oxyethyl]dipyrido[3,2-[1,4]diazepin-6-one, HIV Reverse transcriptase P51, HIV Reverse transcriptase P66, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N- versus O-alkylation: Utilizing NMR methods to establish reliable primary structure determinations for drug discovery.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3WE8
 
 | | Pim-1 kinase in complex with Ruthenium-based inhibitor | | Descriptor: | Cyclopentadienyl(carbon monoxide)(7-oxo-7,10-dihydro-1,8-phenanthrolin-10-yl-kappa~2~C~10~,N~1~)ruthenium, SULFATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Waehler, K, Kraeling, K, Steuber, H, Meggers, E. | | Deposit date: | 2013-07-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Non-ATP-Mimetic Organometallic Protein Kinase Inhibitor
ChemistryOpen, 2, 2013
|
|
5QU2
 
 | | Crystal Structure of human Nck SH3.1 in complex with peptide PPPVPNPDY | | Descriptor: | ACE-PRO-PRO-PRO-VAL-PRO-ASN-PRO-ASP-TYR-NH2, Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
2AZA
 
 | |
5QU7
 
 | |
