8XFR
 
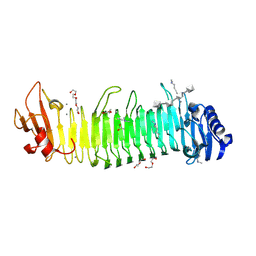 | |
8XFQ
 
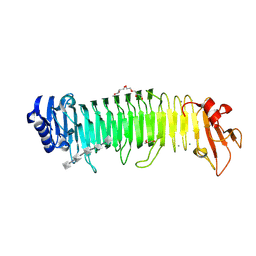 | |
6SQC
 
 | | Crystal structure of complex between nuclear coactivator binding domain of CBP and [1040-1086]ACTR containing alpha-methylated Leu1055 and Leu1076 | | Descriptor: | 1,2-ETHANEDIOL, Maltose/maltodextrin-binding periplasmic protein,CREB-binding protein, Nuclear receptor coactivator 3, ... | | Authors: | Bauer, V, Schmidtgall, B, Gogl, G, Dolenc, j, Osz, J, Kostmann, C, Mitschler, A, Cousido-Siah, A, Rochel, N, Trave, G, Kieffer, B, Torbeev, V. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational editing of intrinsically disordered protein by alpha-methylation.
Chem Sci, 12, 2020
|
|
6SMV
 
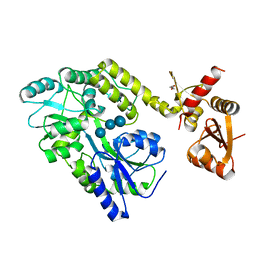 | | Structure of HPV49 E6 protein in complex with MAML1 LxxLL motif | | Descriptor: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Mastermind-like protein 1, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6SIV
 
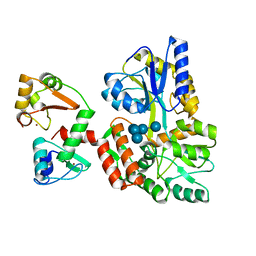 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
8J2P
 
 | | Crystal structure of PML B-box2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
6SZH
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8JBH
 
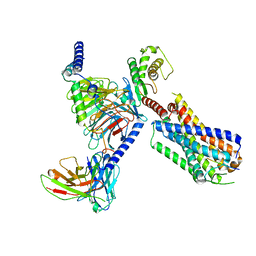 | | Substance P bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBG
 
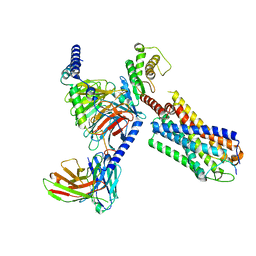 | | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8IZJ
 
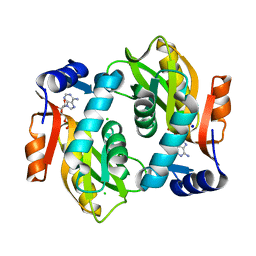 | |
4R0Y
 
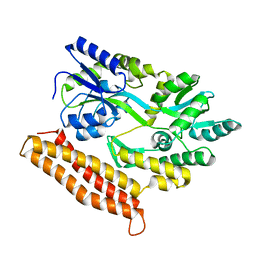 | |
4RG5
 
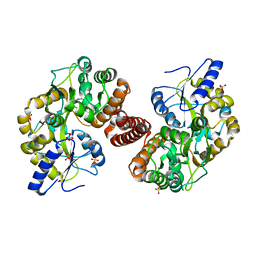 | | Crystal Structure of S. Pombe SMN YG-Dimer | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein, Survival Motor Neuron protein chimera, ... | | Authors: | Gupta, K, Martin, R.S, Sarachan, K.L, Sharp, B, Van Duyne, G.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomeric Properties of Survival Motor NeuronGemin2 Complexes.
J.Biol.Chem., 290, 2015
|
|
4QVH
 
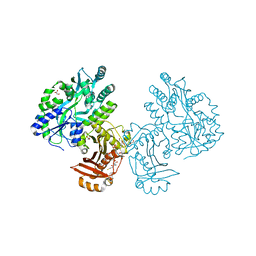 | | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein | | Descriptor: | CITRATE ANION, COENZYME A, GLYCEROL, ... | | Authors: | Jung, J, Bashiri, G, Johnston, J.M, Baker, E.N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein.
J.Struct.Biol., 188, 2014
|
|
4RWF
 
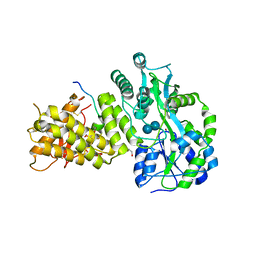 | |
5RJP
 
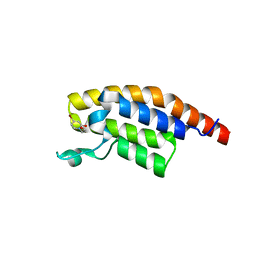 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00024672 | | Descriptor: | 4-bromo-1-(2-hydroxyethyl)pyridin-2(1H)-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK3
 
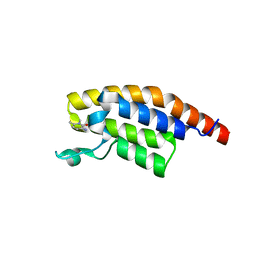 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1501469697 | | Descriptor: | 3-amino-1,6-dimethylpyridin-2(1H)-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKN
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z373768900 | | Descriptor: | N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJW
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z256709556 | | Descriptor: | 3-methylthiophene-2-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKB
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z2004563941 | | Descriptor: | (1S)-1-(1-cyclopentyl-1H-pyrazol-4-yl)ethan-1-ol, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKR
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1432018343 | | Descriptor: | (2S)-2-[(5-chloro-3-fluoropyridin-2-yl)amino]propan-1-ol, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
4TSM
 
 | | MBP-fusion protein of PilA1 from C. difficile R20291 residues 26-166 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein, pilin chimera, ... | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-06-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
5S9E
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | 4-(5-methylfuran-2-carbonyl)-N-[(pyridin-3-yl)methyl]piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5S9C
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | N-(cyclopropylmethyl)-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5S99
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
