1AP4
 
 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-SATURATED STATE, NMR, 40 STRUCTURES | | Descriptor: | CALCIUM ION, CARDIAC N-TROPONIN C | | Authors: | Li, M.X, Spyracopoulos, L, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
1C7W
 
 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
5K58
 
 | |
7XQI
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQJ
 
 | | Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation) | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
6M5Z
 
 | | Catalytic domain of GH30 xylanase C from Talaromyces cellulolyticus | | Descriptor: | ACETATE ION, GH30 Xylanase C, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2020-03-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of GH30-7 endoxylanase C from the filamentous fungus Talaromyces cellulolyticus.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8AV0
 
 | | small molecule stabilizer (compound 1) for C-RAF pS259 and 14-3-3 | | Descriptor: | 1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-4-carboxamide, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6MFT
 
 | | Crystal structure of glycosylated 426c HIV-1 gp120 core G459C in complex with glVRC01 A60C heavy chain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weidle, C, Pancera, M, Stamatatos, L, Gray, M. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
8B78
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{R})-8-(2-chloranyl-6-oxidanyl-phenyl)-7-fluoranyl-9-prop-1-ynyl-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
8B6I
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{S})-7-chloranyl-8-(5-methyl-2~{H}-indazol-4-yl)-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
7O6N
 
 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
6YXM
 
 | | Crystal structure of ACPA 1F2 in complex with CII-C-39-CIT | | Descriptor: | ACPA 1F2 Fab fragment - heavy chain, ACPA 1F2 Fab fragment - light chain, CII-C-39-CIT, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
2W7X
 
 | | Cellular inhibition of checkpoint kinase 2 and potentiation of cytotoxic drugs by novel Chk2 inhibitor PV1019 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[4-[(E)-N-carbamimidamido-C-methyl-carbonimidoyl]phenyl]-7-nitro-1H-indole-2-carboxamide, ... | | Authors: | Jobson, A.G, Lountos, G.T, Lorenzi, P.L, Llamas, J, Connelly, J, Tropea, J.E, Onda, A, Kondapaka, S, Zhang, G, Caplen, N.J, Caredellina, J.H, Monks, A, Self, C, Waugh, D.S, Shoemaker, R.H, Pommier, Y. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cellular Inhibition of Chk2 Kinase and Potentiation of Camptothecins and Radiation by the Novel Chk2 Inhibitor Pv1019.
J.Pharmacol.Exp.Ther., 331, 2009
|
|
6ZDB
 
 | | NMR structural analysis of yeast Cox13 reveals its C-terminus in interaction with ATP | | Descriptor: | Cytochrome c oxidase subunit 13, mitochondrial | | Authors: | Shu, Z, Pontus, P, Peter, B, Lena, M, Pia, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the yeast cytochrome c oxidase subunit Cox13 and its interaction with ATP.
Bmc Biol., 19, 2021
|
|
6R8O
 
 | | Human Cyclophilin D in complex with 1-(((2R,3S,6R)-3-hydroxy-2,3,4,6-tetrahydro-1H-2,6-methanobenzo[c][1,5]oxazocin-8-yl)methyl)-3-(2-((R)-2-(2-(methylthio)phenyl)pyrrolidin-1-yl)-2-oxoethyl)urea | | Descriptor: | 1-(((2R,3S,6R)-3-hydroxy-2,3,4,6-tetrahydro-1H-2,6-methanobenzo[c][1,5]oxazocin-8-yl)methyl)-3-(2-((R)-2-(2-(methylthio)phenyl)pyrrolidin-1-yl)-2-oxoethyl)urea, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5V61
 
 | | Phospho-ERK2 bound to bivalent inhibitor SBP2 | | Descriptor: | 2-oxo-6,9,12,15-tetraoxa-3-azaoctadecan-18-oic acid, 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4XQ0
 
 | |
8QBU
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR CX-4945 AND THE ALPHA-D-POCKET LIGAND 3,4-DICHLORO PHENETHYLAMINE (DPA) | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, ... | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
7Q6Z
 
 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to IMP-1575 | | Descriptor: | 2-(2-methylpropylamino)-1-[(4R)-4-(6-methylpyridin-2-yl)-6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl]ethanone, CHOLESTEROL, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
5V62
 
 | | Phospho-ERK2 bound to bivalent inhibitor SBP3 | | Descriptor: | 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4XPZ
 
 | |
6PSF
 
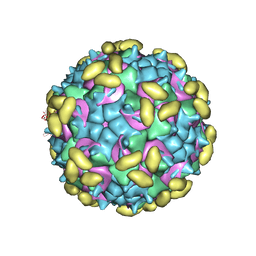 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SXG
 
 | |
8SXE
 
 | |
8SXF
 
 | |
