3B4Y
 
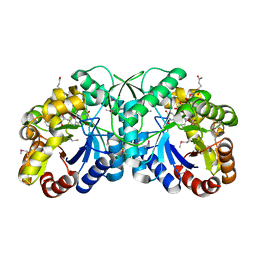 | | FGD1 (Rv0407) from Mycobacterium tuberculosis | | Descriptor: | CITRATE ANION, COENZYME F420, PROBABLE F420-DEPENDENT GLUCOSE-6-PHOSPHATE DEHYDROGENASE FGD1 | | Authors: | Bashiri, G, Squire, C.J, Moreland, N.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-25 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding
J.Biol.Chem., 283, 2008
|
|
4OIM
 
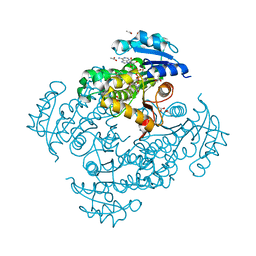 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
2CJD
 
 | |
2CJG
 
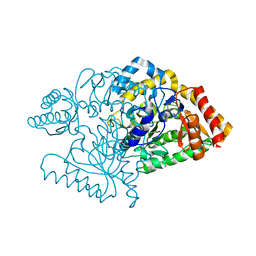 | | Lysine aminotransferase from M. tuberculosis in bound PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-LYSINE-EPSILON AMINOTRANSFERASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2006-04-01 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Direct Evidence for a Glutamate Switch Necessary for Substrate Recognition: Crystal Structures of Lysine Epsilon-Aminotransferase (Rv3290C) from Mycobacterium Tuberculosis H37Rv.
J.Mol.Biol., 362, 2006
|
|
2KNG
 
 | | Solution structure of C-domain of Lsr2 | | Descriptor: | Protein lsr2 | | Authors: | Li, Y, Xia, B. | | Deposit date: | 2009-08-22 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 2010
|
|
2ISZ
 
 | | Crystal structure of a two-domain IdeR-DNA complex crystal form I | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
3IIQ
 
 | |
3IHG
 
 | | Crystal structure of a ternary complex of aklavinone-11 hydroxylase with FAD and aklavinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RdmE, SULFATE ION, ... | | Authors: | Lindqvist, Y, Koskiniemi, H, Jansson, A, Sandalova, T, Schneider, G. | | Deposit date: | 2009-07-30 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
J.Mol.Biol., 393, 2009
|
|
2ISY
 
 | | Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR) | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2UXS
 
 | | 2.7A crystal structure of inorganic pyrophosphatase (Rv3628) from Mycobacterium tuberculosis at pH 7.5 | | Descriptor: | INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Cole, R.E, Cianci, M, Hall, J.F, Matsuda, T, Kigawa, T, Yokoyama, S, Hasnain, S.S, Tabernero, L. | | Deposit date: | 2007-03-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Rv3628: An Inorganic Pyrophosphatase from Mycobacterium Tuberculosis
To be Published
|
|
2IT0
 
 | | Crystal structure of a two-domain IdeR-DNA complex crystal form II | | Descriptor: | ACETATE ION, Iron-dependent repressor ideR, NICKEL (II) ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
3DWZ
 
 | | Meropenem Covalent Adduct with TB Beta-lactamase | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Hugonnet, J.E, Blanchard, J.S. | | Deposit date: | 2008-07-23 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis
Science, 323, 2009
|
|
3ZS4
 
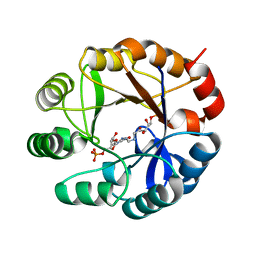 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND PRFAR | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, PHOSPHORIC ACID MONO-[5-({[5-CARBAMOYL-3-(5-PHOSPHONOOXY-5-DEOXY-RIBOFURANOSYL)- 3H-IMIDAZOL-4-YLAMINO]-METHYL}-AMINO)-2,3,4-TRIHYDROXY-PENTYL] ESTER | | Authors: | Due, A.V, Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Phosphoribosyl Isomerase with Bound Prfar
To be Published
|
|
4ADL
 
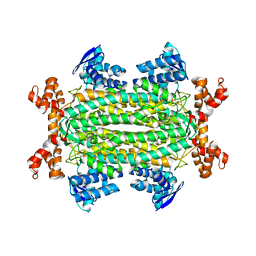 | | Crystal structures of Rv1098c in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FUMARATE HYDRATASE CLASS II | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
4ADM
 
 | | Crystal structure of Rv1098c in complex with meso-tartrate | | Descriptor: | FUMARATE HYDRATASE CLASS II, GLYCEROL, S,R MESO-TARTARIC ACID | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-27 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
2CJH
 
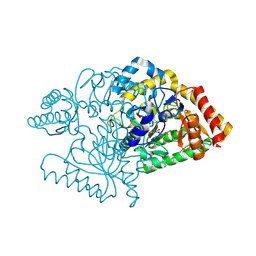 | |
2HYX
 
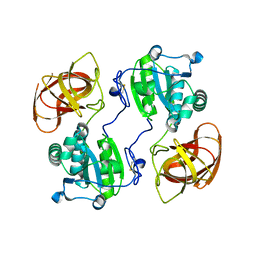 | |
2FHG
 
 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
4B6C
 
 | | Structure of the M. smegmatis GyrB ATPase domain in complex with an aminopyrazinamide | | Descriptor: | 6-(3,4-dimethylphenyl)-3-[[4-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]amino]pyrazine-2-carboxamide, DNA gyrase subunit B,DNA gyrase subunit B,DNA gyrase subunit B, SODIUM ION | | Authors: | Tucker, J.A, Shirude, P.S, Madhavapeddi, P, Hussein, S, Basu, R, Ghorpade, S. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazinamides: novel and specific GyrB inhibitors that kill replicating and nonreplicating Mycobacterium tuberculosis.
ACS Chem. Biol., 8, 2013
|
|
3WO5
 
 | | Crystal structure of S147Q of Rv2613c from Mycobacterium tuberculosis | | Descriptor: | AP-4-A phosphorylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mori, S, Wachino, J, Arakawa, Y, Shibayama, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Role of Ser-147 and Ala-149 in catalytic activity of diadenosine tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4BQR
 
 | | Mtb InhA complex with Methyl-thiazole compound 11 | | Descriptor: | (NZ)-2-[2,6-bis(fluoranyl)phenyl]-N-[5-[(1S)-1-(4-methyl-1,3-thiazol-2-yl)-1-oxidanyl-ethyl]-3H-1,3,4-thiadiazol-2-ylidene]ethanamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
4ECP
 
 | |
8QVO
 
 | | L211Q, L254N, T262G mutant of carboxypeptidase T from Thermoactinomyces vulgaris | | Descriptor: | CALCIUM ION, Carboxypeptidase T, SULFATE ION, ... | | Authors: | Timofeev, V.I, Dorovatovskii, P.V, Lazarenko, V.A, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2023-10-18 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | L211Q, L254N, T262G mutant of carboxypeptidase T from Thermoactinomyces vulgaris
To Be Published
|
|
2E00
 
 | |
