2BCY
 
 | | Crystal Structure of a minimal, mutant all-RNA hairpin ribozyme (U39C, G8MTU) | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*(MTU)P*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2005-10-19 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
5DV3
 
 | |
1BL6
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
7EWJ
 
 | | Toxin-antitoxin complex from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PemI inhibitor, ... | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
1PSW
 
 | | Structure of E. coli ADP-heptose lps heptosyltransferase II | | Descriptor: | ADP-HEPTOSE LPS HEPTOSYLTRANSFERASE II | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of E. coli ADP-heptose lps heptosyltransferase II
To be Published
|
|
7ZE1
 
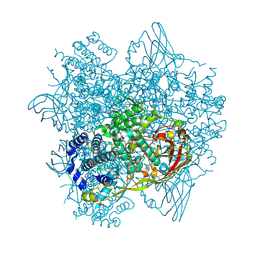 | | Tribolium castaneum hexamerin 2 | | Descriptor: | Larval serum protein 1 gamma chain-like Protein | | Authors: | Valentova, L, Fuzik, T, Plevka, P. | | Deposit date: | 2022-03-30 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Polyelectrolyte coating of cryo-EM grids improves lateral distribution and prevents aggregation of macromolecules.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1SJS
 
 | |
6GU6
 
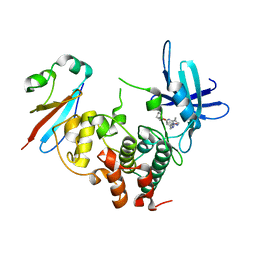 | | CDK1/Cks2 in complex with Dinaciclib | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
1S39
 
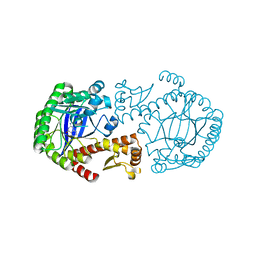 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-aminoquinazolin-4(3H)-one | | Descriptor: | 2-AMINOQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Brenk, R, Meyer, E.A, Reuter, K, Garcia, G.A, Stubbs, M.T, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and In Vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-Based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
6GU4
 
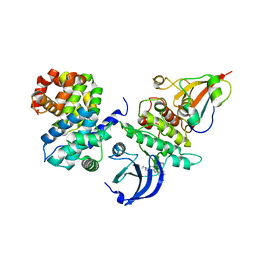 | | CDK1/CyclinB/Cks2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
1SBQ
 
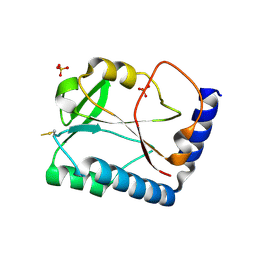 | | Crystal Structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae at 2.2 resolution | | Descriptor: | 5,10-Methenyltetrahydrofolate synthetase homolog, SULFATE ION | | Authors: | Chen, S, Shin, D.H, Pufan, R, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-02-10 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
2CA1
 
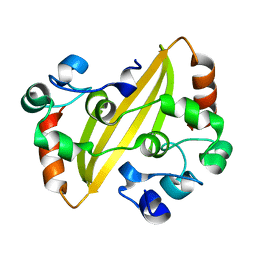 | | Crystal structure of the IBV coronavirus nucleocapsid | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collison, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
7F80
 
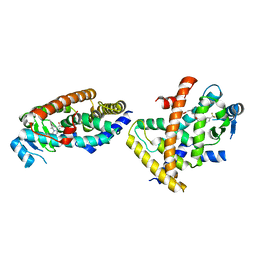 | |
7C9I
 
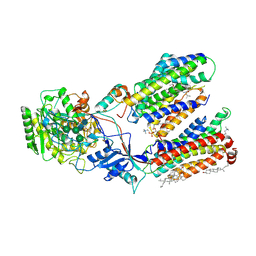 | | Human gamma-secretase in complex with small molecule L-685,458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
2B57
 
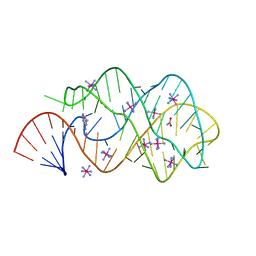 | | Guanine Riboswitch C74U mutant bound to 2,6-diaminopurine | | Descriptor: | 65-MER, 9H-PURINE-2,6-DIAMINE, ACETATE ION, ... | | Authors: | Gilbert, S.D, Stoddard, C.D, Wise, S.J, Batey, R.T. | | Deposit date: | 2005-09-27 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain.
J.Mol.Biol., 359, 2006
|
|
1T77
 
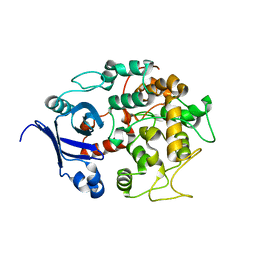 | | Crystal structure of the PH-BEACH domains of human LRBA/BGL | | Descriptor: | Lipopolysaccharide-responsive and beige-like anchor protein | | Authors: | Gebauer, D, Li, J, Jogl, G, Shen, Y, Myszka, D.G, Tong, L. | | Deposit date: | 2004-05-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the PH-BEACH Domains of Human LRBA/BGL
Biochemistry, 43, 2004
|
|
2CMO
 
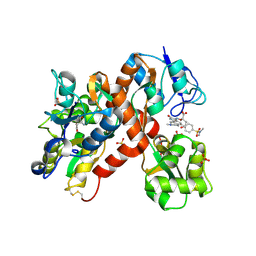 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | Descriptor: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
5DV6
 
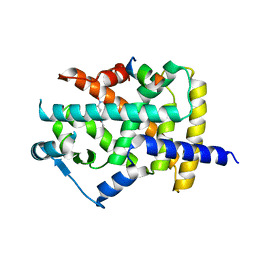 | |
5DWL
 
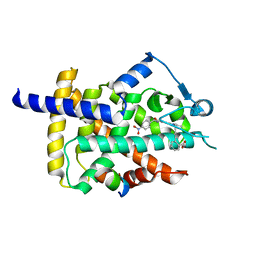 | | Human PPARgamma ligand binding dmain in complex with SR1664 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1S)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y. | | Deposit date: | 2015-09-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human PPARgamma ligand binding dmain in complex with SR1664
To Be Published
|
|
1T8O
 
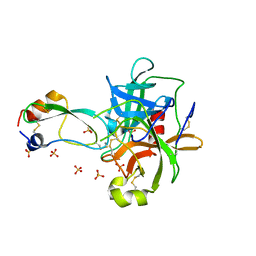 | | CRYSTAL STRUCTURE OF THE P1 TRP BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-13 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
7CUO
 
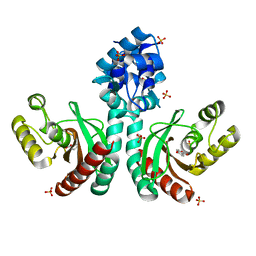 | | IclR transcription factor complexed with 4-hydroxybenzoic acid from Microbacterium hydrocarbonoxydans | | Descriptor: | P-HYDROXYBENZOIC ACID, SULFATE ION, Transcription factor | | Authors: | Akiyama, T, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-08-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
5DV8
 
 | | Human PPARgamma ligand binding dmain complexed with SB1451 in a covalent bonded form | | Descriptor: | N-[2-({2-[({4-[(4-methylpiperazin-1-yl)methyl]benzoyl}amino)methyl]benzyl}oxy)phenyl]-3-nitrobenzamide, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y. | | Deposit date: | 2015-09-21 | | Release date: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Human PPARgamma ligand binding dmain complexed with SB1451 in a covalent bonded form
To Be Published
|
|
1QF6
 
 | | STRUCTURE OF E. COLI THREONYL-TRNA SYNTHETASE COMPLEXED WITH ITS COGNATE TRNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, THREONINE TRNA, THREONYL-TRNA SYNTHETASE, ... | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 1999-04-06 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site
Cell(Cambridge,Mass.), 97, 1999
|
|
1Z3L
 
 | | X-Ray Crystal Structure of a Mutant Ribonuclease S (F8Anb) | | Descriptor: | Ribonuclease pancreatic, S-Peptide, S-Protein, ... | | Authors: | Das, M, Vasudeva Rao, B, Ghosh, S, Varadarajan, R. | | Deposit date: | 2005-03-14 | | Release date: | 2005-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Attempts to delineate the relative contributions of changes in hydrophobicity and packing to changes in stability of ribonuclease S mutants.
Biochemistry, 44, 2005
|
|
1Z9D
 
 | | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes | | Descriptor: | SULFATE ION, uridylate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes
To be Published
|
|
