7Z43
 
 | |
6BRH
 
 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
6BRK
 
 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
2YFD
 
 | | STRUCTURAL AND FUNCTIONAL INSIGHTS OF DR2231 PROTEIN, THE MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE FROM DEINOCOCCUS RADIODURANS, COMPLEXED WITH Mg and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goncalves, A.M.D, De Sanctis, D, Mcsweeney, S.M. | | Deposit date: | 2011-04-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
3HKN
 
 | | Human carbonic anhydrase II in complex with (2,3,4,6-Tetra-O-acetyl-beta-D-galactopyranosyl) -(1-4)-1,2,3,6-tetra-O-acetyl-1-thio-beta-D-glucopyranosylsulfonamide | | Descriptor: | 2,3,4,6-tetra-O-acetyl-beta-D-galactopyranose-(1-4)-(1S)-2,3,6-tri-O-acetyl-1,5-anhydro-1-sulfamoyl-D-glucitol, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
3VU4
 
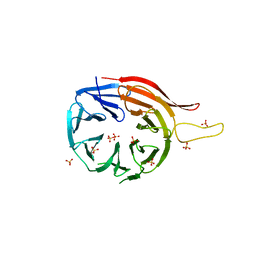 | |
3VZ8
 
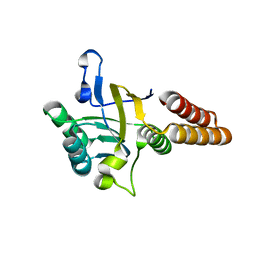 | |
3HKQ
 
 | | Human carbonic anhydrase II in complex with 1-S-D-Galactopyranosylsulfonamide | | Descriptor: | (1S)-1,5-anhydro-1-sulfamoyl-D-galactitol, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
4PLU
 
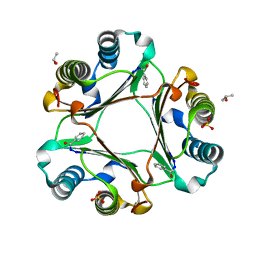 | |
4PKZ
 
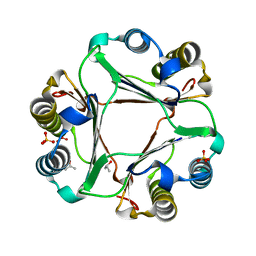 | |
1MFF
 
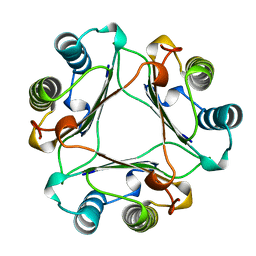 | | MACROPHAGE MIGRATION INHIBITORY FACTOR Y95F MUTANT | | Descriptor: | MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Taylor, A.B, Stamps, S.L, Wang, S.C, Hackert, M.L, Whitman, C.P. | | Deposit date: | 1998-10-19 | | Release date: | 1999-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of the phenylpyruvate tautomerase activity of macrophage migration inhibitory factor: properties of the P1G, P1A, Y95F, and N97A mutants.
Biochemistry, 39, 2000
|
|
3VGO
 
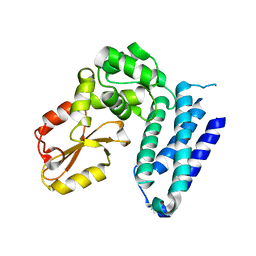 | |
2LQ8
 
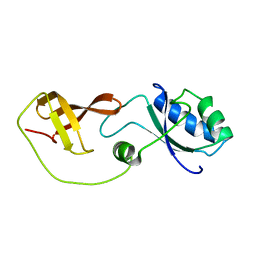 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
6STY
 
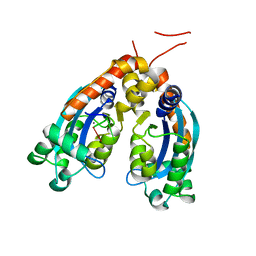 | | Human REXO2 exonuclease in complex with RNA. | | Descriptor: | CALCIUM ION, Oligoribonuclease, mitochondrial, ... | | Authors: | Malik, D, Szewczyk, M, Szczesny, R, Nowotny, M. | | Deposit date: | 2019-09-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Human REXO2 controls short mitochondrial RNAs generated by mtRNA processing and decay machinery to prevent accumulation of double-stranded RNA.
Nucleic Acids Res., 48, 2020
|
|
2LHJ
 
 | |
3IL9
 
 | | Structure of E. coli FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
6C1I
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with T0070907 | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Fuhrmann, J, Brust, R, Kojetin, D.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A structural mechanism for directing corepressor-selective inverse agonism of PPAR gamma.
Nat Commun, 9, 2018
|
|
3IN7
 
 | |
3VX7
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3IB9
 
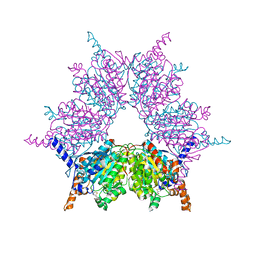 | | Propionyl-CoA Carboxylase Beta Subunit, D422L | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase complex B subunit, SULFATE ION | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3ID9
 
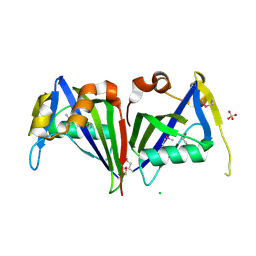 | |
1MIF
 
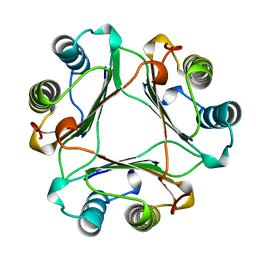 | | MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) | | Descriptor: | MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Sun, H.-W, Lolis, E. | | Deposit date: | 1996-01-26 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2YF4
 
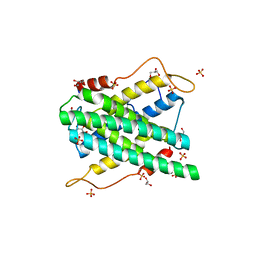 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, Apo structure | | Descriptor: | GLYCEROL, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, SULFATE ION | | Authors: | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
3IA3
 
 | |
3UKM
 
 | |
