4Q1L
 
 | | Crystal structure of Leucurolysin-a complexed with an endogenous tripeptide (QSW). | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ferreira, R.N, Sanchez, E.O.F, Pimenta, A.M.C, Nagem, R.A.P. | | Deposit date: | 2014-04-03 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Leucurolysin-a complexed with an endogenous tripeptide (QSW).
To be Published
|
|
1VDQ
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
to be published
|
|
4HBY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
1VDT
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space
to be published
|
|
3F80
 
 | |
4BLW
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylhomocysteine (AdoHcy) and Adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
2WVJ
 
 | | Mutation of Thr163 to Ser in Human Thymidine Kinase Shifts the Specificity from Thymidine towards the Nucleoside Analogue Azidothymidine | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, CYTOSOLIC, ... | | Authors: | Skovgaard, T, Uhlin, U, Munch-Petersen, B. | | Deposit date: | 2009-10-17 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative Active-Site Mutation Study of Human and Caenorhabditis Elegans Thymidine Kinase 1.
FEBS J., 279, 2012
|
|
1QUD
 
 | | L99G MUTANT OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1VJO
 
 | |
5Z09
 
 | | ST0452(Y97N)-UTP binding form | | Descriptor: | Dual sugar-1-phosphate nucleotidylyltransferase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
3F67
 
 | | Crystal Structure of Putative Dienelactone Hydrolase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Putative Dienelactone Hydrolase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
1AJ8
 
 | | CITRATE SYNTHASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Ferguson, J.M.C, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution,.
Biochemistry, 36, 1997
|
|
1ALC
 
 | |
1VP2
 
 | |
1VKI
 
 | |
7FRY
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 6 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{2-[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]ethyl}-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
1HXW
 
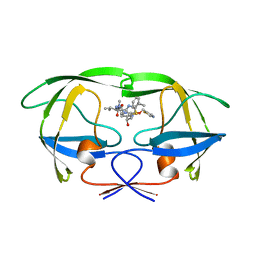 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-84538 | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Park, C.H, Nienaber, V, Kong, X.P. | | Deposit date: | 1997-01-24 | | Release date: | 1998-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7FS7
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 20 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-N-(3,4-dihydroxyphenyl)-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRZ
 
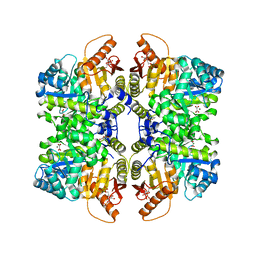 | | Structure of liver pyruvate kinase in complex with allosteric modulator 7 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3,4-dihydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRX
 
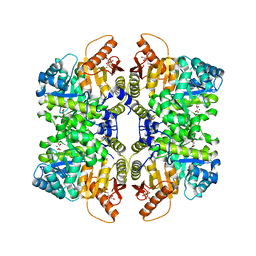 | | Structure of liver pyruvate kinase in complex with allosteric modulator 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
3UTS
 
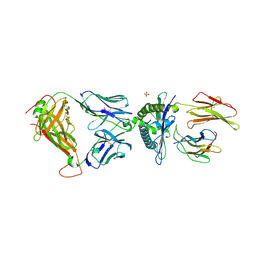 | | 1E6-A*0201-ALWGPDPAAA Complex, Monoclinic | | Descriptor: | 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
7FS0
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 8 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3,4-dihydroxy-N-{2-[4-(3-hydroxybenzene-1-sulfonyl)phenyl]ethyl}benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
1VQT
 
 | |
1HUN
 
 | |
7FS4
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 16 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4,4',5-trihydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}[1,1'-biphenyl]-2-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
