7JJA
 
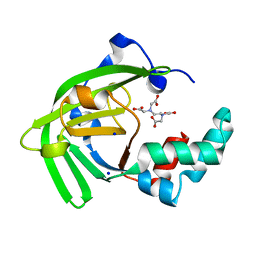 | | Crystal structure of the ZinT-like domain of Streptococcus pneumoniae AdcA in the apo form | | Descriptor: | SODIUM ION, Zinc-binding lipoprotein AdcA, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7JJM
 
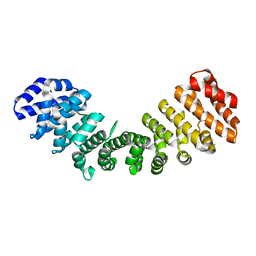 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS | | Descriptor: | CHLORIDE ION, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McGuaig, R, Tan, H.Y.A, Hardy, C, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
7JJL
 
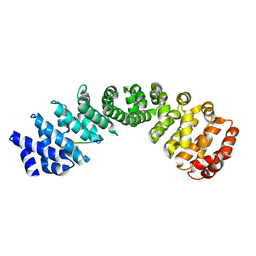 | | Crystal structure of Importin Alpha 3 in complex with human LSD1 NLS | | Descriptor: | Importin subunit alpha-3, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, H.Y.A, Hardy, K, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
7JK7
 
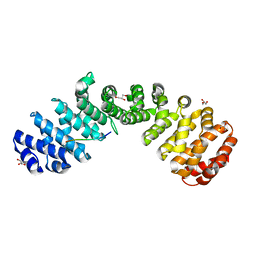 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS S111E mutant | | Descriptor: | GLYCEROL, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, A.H.Y, Hardy, K, Seddiki, N, Ali, S, Dahlstron, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
7JHM
 
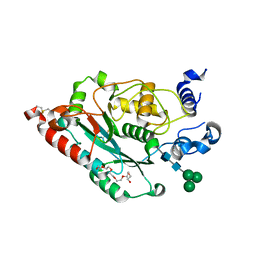 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7EUA
 
 | | X-ray structure of a P93A Monellin mutant | | Descriptor: | Monellin, SULFATE ION | | Authors: | Manjula, R, Bhatia, S, Jayant, B, Ramaswamy, S, Gosavi, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | X-ray structure of a P93A Monellin mutant
To Be Published
|
|
7FE5
 
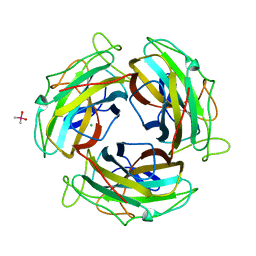 | |
7FE6
 
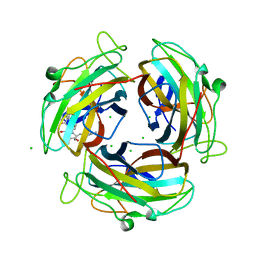 | |
7FE0
 
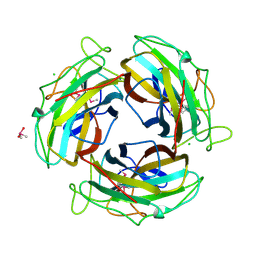 | |
7FG6
 
 | |
7F2R
 
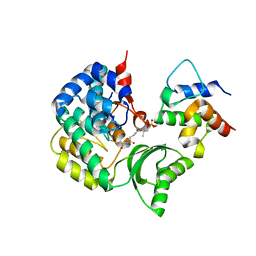 | | Crystal structure of VinK-VinL covalent complex formed with a pantetheineamide cross-linking probe | | Descriptor: | Acyl-carrier-protein, Malonyl-CoA-[acyl-carrier-protein] transacylase, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Miyanaga, A, Ouchi, R, Kudo, F, Eguchi, T. | | Deposit date: | 2021-06-14 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex structure of the acyltransferase VinK and the carrier protein VinL with a pantetheine cross-linking probe.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7FFP
 
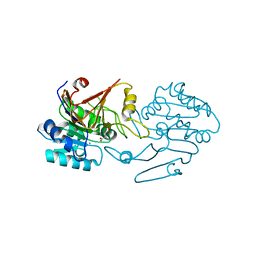 | | Crystal structure of di-peptidase-E from Xenopus laevis | | Descriptor: | ASPARTIC ACID, Alpha-aspartyl dipeptidase, CALCIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
7F26
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Liang, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7EQL
 
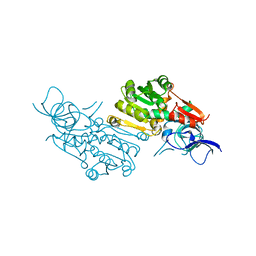 | | Crystal structure of (+)-pulegone reductase from Mentha piperita | | Descriptor: | (+)-pulegone reductase | | Authors: | Lin, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Functional Characterization and Structural Insights Into Stereoselectivity of Pulegone Reductase in Menthol Biosynthesis.
Front Plant Sci, 12, 2021
|
|
7ESW
 
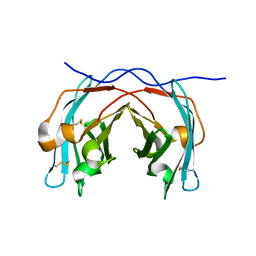 | |
7F9X
 
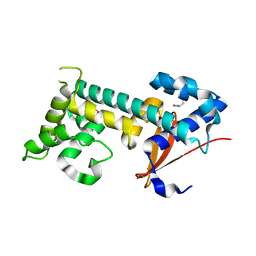 | | Structure of the second OTU domain of LotA | | Descriptor: | LotA | | Authors: | Iwai, T, Takekawa, T, Kubori, T, Nagai, H, Imada, K. | | Deposit date: | 2021-07-05 | | Release date: | 2021-10-20 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Ubiquitin Recognition by a Bacterial Ovarian Tumor Deubiquitinase LotA.
J.Bacteriol., 204, 2022
|
|
7GE4
 
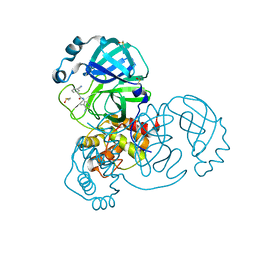 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714a760b-19 (Mpro-x11318) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-amino-4-methylpyridin-3-yl)-2-(3-cyanophenyl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GEZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6747fa38-1 (Mpro-x11541) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GEF
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-93268d01-11 (Mpro-x11428) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-(piperidin-1-yl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GF5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-7 (Mpro-x11562) | | Descriptor: | 2-(2-butoxy-5-chlorophenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GEM
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-15 (Mpro-x11485) | | Descriptor: | 2-(3-chlorophenyl)-N-(4-methylpyridazin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GEW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-bb423b95-2 (Mpro-x11530) | | Descriptor: | 2-(3-chlorophenyl)-N-(1,6-naphthyridin-8-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GF7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TAT-ENA-80bfd3e5-7 (Mpro-x11579) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-1,5-dicyano-4-(methylsulfanyl)-3-azaspiro[5.5]undeca-2,4-dien-2-yl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GF3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-5 (Mpro-x11557) | | Descriptor: | (2S)-2-(3-chlorophenyl)-3-hydroxy-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GFG
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with CHO-MSK-6e55470f-5 (Mpro-x11723) | | Descriptor: | 2-{3-chloro-5-[(3-methyl-1,2,4-oxadiazol-5-yl)methoxy]phenyl}-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
