8ORA
 
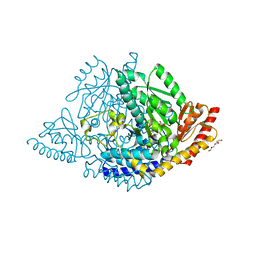 | | Human holo aromatic L-amino acid decarboxylase (AADC) external aldimine with L-Dopa methylester | | Descriptor: | Dopa decarboxylase (Aromatic L-amino acid decarboxylase), PYRIDOXAL-5'-PHOSPHATE, TETRAETHYLENE GLYCOL, ... | | Authors: | Bisello, G, Perduca, M, Bertoldi, M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: Implication in aromatic amino acid decarboxylase deficiency.
Protein Sci., 32, 2023
|
|
8ONJ
 
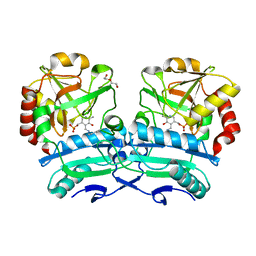 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | Descriptor: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
8BLT
 
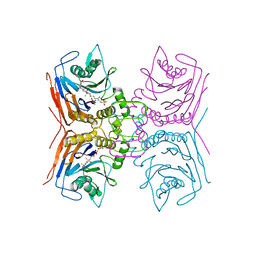 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with taurocholate (TCA) | | Descriptor: | Bile salt hydrolase, TAUROCHOLIC ACID | | Authors: | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
8BLS
 
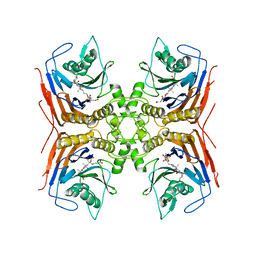 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with Glycocholate (GCA) | | Descriptor: | Bile salt hydrolase, GLYCOCHOLIC ACID | | Authors: | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
2BG2
 
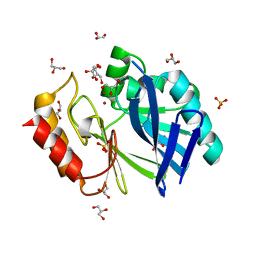 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in the buffer. 1mM DTT and 1mM TCEP- HCl were used as reducing agents. Cys221 is reduced. | | Descriptor: | BETA-LACTAMASE II, CHLORIDE ION, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism.
Biochemistry, 44, 2005
|
|
1HXP
 
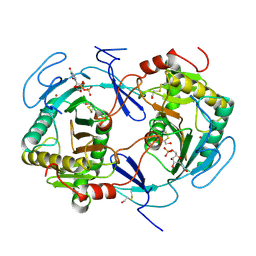 | | NUCLEOTIDE TRANSFERASE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1995-06-09 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
8W8O
 
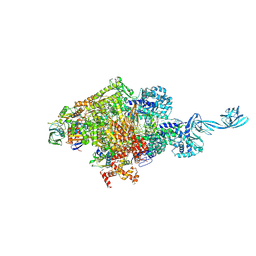 | |
7U22
 
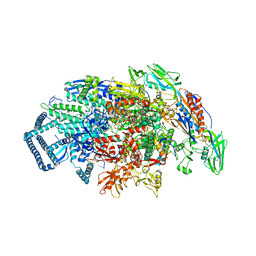 | | Mycobacterium tuberculosis RNA polymerase sigma A holoenzyme open promoter complex containing UMN-7 | | Descriptor: | (9S,14S,15R,16S,17R,18R,19R,20S,21S,25R)-5,6,18,20-tetrahydroxy-14-methoxy-7,9,15,17,19,21,25-heptamethyl-1'-(2-methylpropyl)-10,26-dioxo-1,3,9,10-tetrahydrospiro[9,4-(epoxypentadecanoimino)furo[2',3':7,8]naphtho[1,2-d]imidazole-2,4'-piperidin]-16-yl benzoate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Redesign of Rifamycin Antibiotics to Overcome ADP-Ribosylation-Mediated Resistance.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8W8P
 
 | |
6GYA
 
 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8W8N
 
 | |
3VIG
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with 1-deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-glucosidase, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7S4A
 
 | | MRG15 complex with PALB2 peptide | | Descriptor: | Mortality factor 4-like protein 1, Partner and localizer of BRCA2 | | Authors: | Korolev, S, Deveryshetty, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structural Insight into the Mechanism of PALB2 Interaction with MRG15.
Genes (Basel), 12, 2021
|
|
3V7X
 
 | | Complex of human carbonic anhydrase II with N-[2-(3,4-dimethoxyphenyl)ethyl]-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, MERCURIBENZOIC ACID, ... | | Authors: | Mader, P, Brynda, J, Rezacova, P. | | Deposit date: | 2011-12-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
5OAW
 
 | |
8ONN
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
2BFZ
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
2XPC
 
 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
8OR9
 
 | |
8RUC
 
 | | ACTIVATED SPINACH RUBISCO COMPLEXED WITH 2-CARBOXYARABINITOL BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Andersson, I, Knight, S, Branden, C.-I. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate.
J.Mol.Biol., 259, 1996
|
|
8D6Y
 
 | |
1HLD
 
 | | STRUCTURES OF HORSE LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH NAD+ AND SUBSTITUTED BENZYL ALCOHOLS | | Descriptor: | 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Ramaswamy, S, Eklund, H, Plapp, B.V. | | Deposit date: | 1993-11-23 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of horse liver alcohol dehydrogenase complexed with NAD+ and substituted benzyl alcohols.
Biochemistry, 33, 1994
|
|
8D6X
 
 | |
