3FSS
 
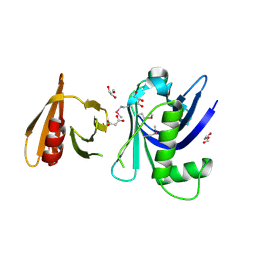 | | Structure of the tandem PH domains of Rtt106 | | Descriptor: | GLYCEROL, Histone chaperone RTT106, MALONIC ACID | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2009-01-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
7LBK
 
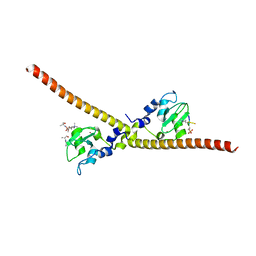 | |
7LBQ
 
 | |
7LBO
 
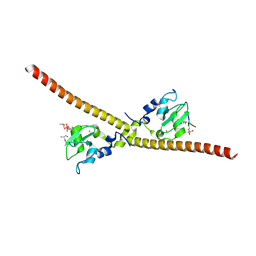 | |
1WZ1
 
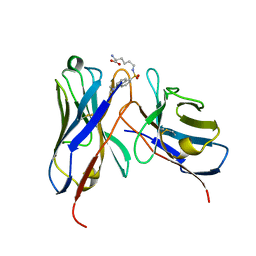 | | Crystal structure of the Fv fragment complexed with dansyl-lysine | | Descriptor: | Ig heavy chain, Ig light chain, N~6~-{[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}-L-LYSINE | | Authors: | Nakasako, M, Oka, T, Mashumo, M, Takahashi, H, Shimada, I, Yamaguchi, Y, Kato, K, Arata, Y. | | Deposit date: | 2005-02-21 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational dynamics of complementarity-determining region H3 of an anti-dansyl Fv fragment in the presence of its hapten
J.Mol.Biol., 351, 2005
|
|
3A1B
 
 | | Crystal structure of the DNMT3A ADD domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, Histone H3.1, ... | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
7KLO
 
 | |
7Y5V
 
 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5U
 
 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
6WW3
 
 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
6F6D
 
 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-12-05 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81810677 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
8Q1H
 
 | | LSD1 Y391K-CoREST bound to Histone H3 N-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 21, 2025
|
|
8Q1G
 
 | | LSD1-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 21, 2025
|
|
8Q1J
 
 | | LSD1 Y391K-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 21, 2025
|
|
8G8G
 
 | | Interaction of H3 tail in LIN28B nucleosome with Oct4 | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
2L43
 
 | | Structural basis for histone code recognition by BRPF2-PHD1 finger | | Descriptor: | Histone H3.3,LINKER,Bromodomain-containing protein 1, ZINC ION | | Authors: | Qin, S, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of unmodified histone H3 by the first PHD finger of Bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases MOZ and MORF
To be Published
|
|
6BHG
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHI
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6BHE
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
3W98
 
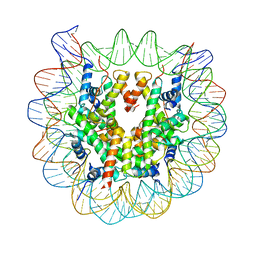 | | Crystal Structure of Human Nucleosome Core Particle lacking H3.1 N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
8P89
 
 | |
8P88
 
 | |
5KDM
 
 | |
6BHH
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
