6JGN
 
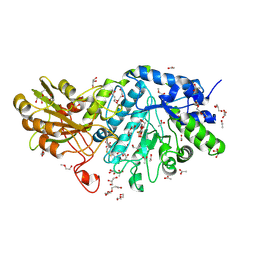 | | Crystal structure of barley exohydrolaseI W434H in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JG7
 
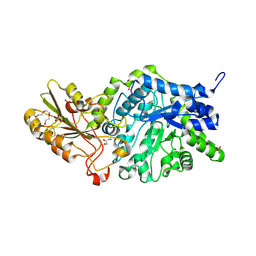 | | Crystal structure of barley exohydrolaseI W286F in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGC
 
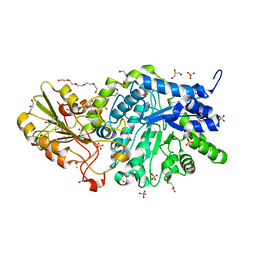 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGS
 
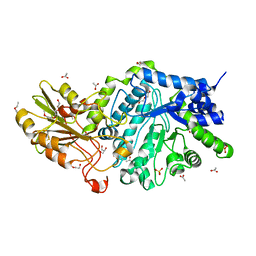 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGK
 
 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JG2
 
 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-nitrophenoxy)-3,5-bis(oxidanyl)oxan-4-yl]sulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGD
 
 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGP
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGA
 
 | | Crystal structure of barley exohydrolaseI W286F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGL
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGO
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6K6V
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGE
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3LPE
 
 | | Crystal structure of Spt4/5NGN heterodimer complex from Methanococcus jannaschii | | Descriptor: | DNA-directed RNA polymerase subunit E'', Putative transcription antitermination protein nusG, ZINC ION | | Authors: | Hirtreiter, A, Damsma, G.E, Cheung, A.C.M, Klose, D, Grohmann, D, Vojnic, E, Martin, A.C.R, Cramer, P, Werner, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif.
Nucleic Acids Res., 38, 2010
|
|
3P2X
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiaral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
3P33
 
 | | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Hua, Q.X, Wan, Z.L, Huang, K, Hu, S.Q, Phillip, N.F, Jia, W.H, Whittingham, J, Dodson, G.G, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2010-10-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin fibrillation is the Janus face of induced fit. A chiral clamp stabilizes the native state at the expense of activity
To be Published
|
|
4D2G
 
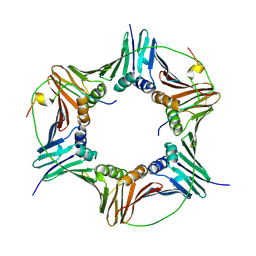 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
5HXY
 
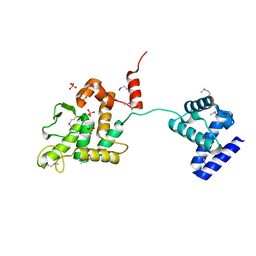 | | Crystal structure of XerA recombinase | | Descriptor: | PHOSPHATE ION, Tyrosine recombinase XerA | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
3V19
 
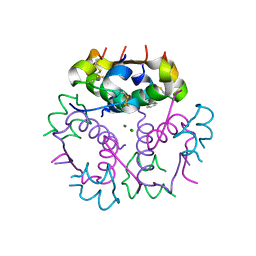 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
6TNG
 
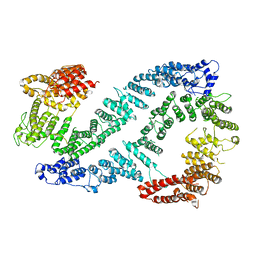 | | Structure of FANCD2 in complex with FANCI | | Descriptor: | Fanconi anemia complementation group I, Uncharacterized protein | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TNF
 
 | | Structure of monoubiquitinated FANCD2 in complex with FANCI and DNA | | Descriptor: | DNA (33-MER), FANCD2, Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TNI
 
 | | Structure of FANCD2 homodimer | | Descriptor: | Uncharacterized protein | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3V1G
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
3ZLJ
 
 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
2DYW
 
 | | A Backbone binding DNA complex | | Descriptor: | (6-AMINOHEXYLAMINE)(TRIAMMINE) PLATINUM(II) COMPLEX, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', SODIUM ION, ... | | Authors: | Komeda, S, Moulaei, T, Woods, K.K, Chikuma, M, Farrell, N.P, Williams, L.D. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A Third Mode of DNA Binding: Phosphate Clamps by a Polynuclear Platinum Complex
J.Am.Chem.Soc., 128, 2006
|
|
