8BZA
 
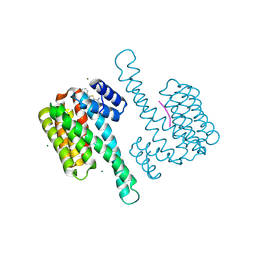 | | single soak stabilizer for ERa - 14-3-3 interaction (AZ555) | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8C4G
 
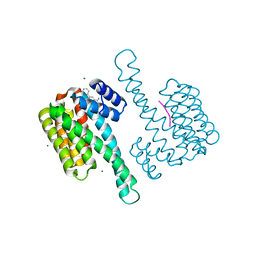 | | Small molecule amidine soak in 14-3-3/ERa (AZ132) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6A6T
 
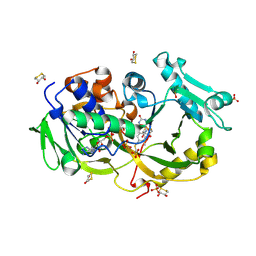 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans with R61G mutation | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
7W9Q
 
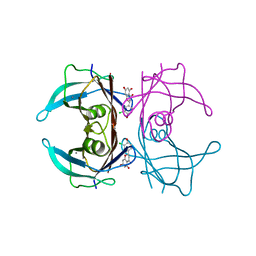 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
5AV3
 
 | |
6ABR
 
 | | Actin interacting protein 5 (Aip5, wild type) | | Descriptor: | Actin binding protein | | Authors: | Sun, J, Xie, Y, Toh, J.D.W, Hong, W, MIao, Y, Gao, Y.G. | | Deposit date: | 2018-07-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
7W9Z
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nitrate | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITRATE ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
5F2P
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 3. | | Descriptor: | 2-(dimethylamino)-6-methyl-pyrido[4,3-d]pyrimidin-5-one, BRD9 | | Authors: | Nar, H, Fiegen, D, Zoephel, A, Bader, G. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
7LYZ
 
 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | Deposit date: | 1977-05-06 | | Release date: | 1977-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
7W9Y
 
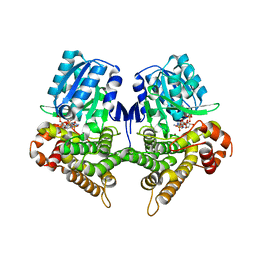 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nickel | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICKEL (II) ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
5BKC
 
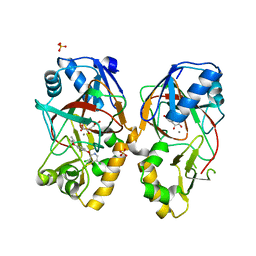 | | Crystal structure of AAD-1 in complex with (R)-diclofop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-{4-[(3,5-dichloropyridin-2-yl)oxy]phenoxy}propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5F4P
 
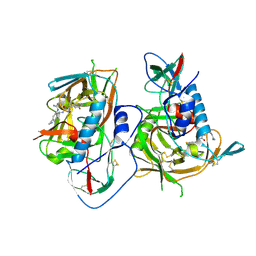 | | HIV-1 gp120 complex with BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
7LWB
 
 | |
7NWK
 
 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
7O2P
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3756 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6A6R
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
7UVD
 
 | | Sco GlgEI-V279S in complex with cyclohexyl carbasugar | | Descriptor: | (1R,4S,5S,6R)-4-(cyclohexylamino)-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Sco GlgEI-V279S in complex with cyclohexyl carbasugar
To Be Published
|
|
5EY6
 
 | |
7CM1
 
 | | Neuraminidase from the Wuhan Asiatic toad influenza virus | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Wang, J. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Lattice-translocation defects in specific crystals of the catalytic head domain of influenza neuraminidase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5F25
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 4. | | Descriptor: | 4-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)benzamide, BRD9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5EY5
 
 | | LBCATS | | Descriptor: | LBCA-b, LBCATS-a, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Busch, F, Rajendran, C, Schlee, S, Heyn, K, Merkl, R, Sterner, R. | | Deposit date: | 2015-11-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | LBCATS
To Be Published
|
|
5BJ4
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 | | Descriptor: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-11 | | Release date: | 2003-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
5C5E
 
 | | Structure of KaiA dimer in complex with C-terminal KaiC peptide at 2.8 A resolution | | Descriptor: | 2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-5-[(sulfanylcarbonyl)amino]benzoic acid, Circadian clock protein KaiA, KaiC C-terminal peptide | | Authors: | Pattanayek, R, Egli, M. | | Deposit date: | 2015-06-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Protein-Protein Interactions in the Cyanobacterial Circadian Clock: Structure of KaiA Dimer in Complex with C-Terminal KaiC Peptides at 2.8 angstrom Resolution.
Biochemistry, 54, 2015
|
|
5F1H
 
 | | Crystal structure of the BRD9 bromodamian in complex with BI-9564. | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
7WRX
 
 | | Structure of Deinococcus radiodurans HerA-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HerA, MAGNESIUM ION | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.40003562 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
