6DRT
 
 | |
2QYU
 
 | | Crystal structure of Salmonella effector protein SopA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Secreted effector protein | | Authors: | Diao, J, Chen, J. | | Deposit date: | 2007-08-15 | | Release date: | 2007-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1VDX
 
 | |
7R71
 
 | | Crystal Structure of the UbArk2C-UbcH5b~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin,E3 ubiquitin-protein ligase RNF165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
6DO5
 
 | | KLHDC2 ubiquitin ligase in complex with USP1 C-end degron | | Descriptor: | Kelch domain-containing protein 2, USP1 C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
6ITS
 
 | |
6DO4
 
 | | KLHDC2 ubiquitin ligase in complex with SelS C-end degron | | Descriptor: | Kelch domain-containing protein 2, SELS C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
5HEG
 
 | |
3QYL
 
 | | Sensitivity of receptor internal motions to ligand binding affinity and kinetic off-rate | | Descriptor: | (7S)-7-methyl-5,6,7,8-tetrahydroquinazoline-2,4-diamine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Collins, E.J, Lee, A.L, Carroll, M.J, Mauldin, R.V, Gromova, A.V, Singleton, S.F. | | Deposit date: | 2011-03-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Evidence for dynamics in proteins as a mechanism for ligand dissociation.
Nat.Chem.Biol., 8, 2012
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
1SHQ
 
 | | Crystal structure of shrimp alkaline phosphatase with magnesium in M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, SULFATE ION, ... | | Authors: | de Backer, M.M.E, McSweeney, S, Lindley, P.F, Hough, E. | | Deposit date: | 2004-02-26 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2EVP
 
 | | The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins | | Descriptor: | BETA-MERCAPTOETHANOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qin, J, Perera, R, Lovelace, L.L, Dawson, J.H, Lebioda, L. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins(,).
Biochemistry, 45, 2006
|
|
5V0I
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, TRYPTOPHAN, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Grimshaw, S.G, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan
To Be Published
|
|
1SHN
 
 | | Crystal structure of shrimp alkaline phosphatase with phosphate bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, SULFATE ION, ... | | Authors: | de Backer, M.M.E, McSweeney, S, Lindley, P.F, Hough, E. | | Deposit date: | 2004-02-26 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-binding and metal-exchange crystallographic studies on shrimp alkaline phosphatase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3QYO
 
 | | Sensitivity of receptor internal motions to ligand binding affinity and kinetic off-rate | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Collins, E.J, Lee, A.L, Carroll, M.J, Mauldin, R.V, Gromova, A.V, Singleton, S.F. | | Deposit date: | 2011-03-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Evidence for dynamics in proteins as a mechanism for ligand dissociation.
Nat.Chem.Biol., 8, 2012
|
|
7ULZ
 
 | |
6A5X
 
 | | FXR-LBD with HNC180 and SRC1 | | Descriptor: | 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
1JTZ
 
 | | CRYSTAL STRUCTURE OF TRANCE/RANKL CYTOKINE. | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 11 | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity
J.Clin.Invest., 108, 2001
|
|
6A5Z
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC180 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
7Z8R
 
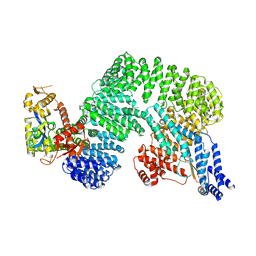 | | CAND1-CUL1-RBX1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
5V3O
 
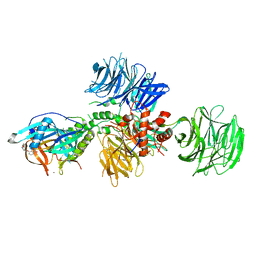 | | Cereblon in complex with DDB1 and CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Matyskiela, M, Pagarigan, B, Chamberlain, P. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos.
J. Med. Chem., 61, 2018
|
|
1UPV
 
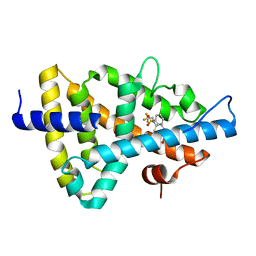 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
1S19
 
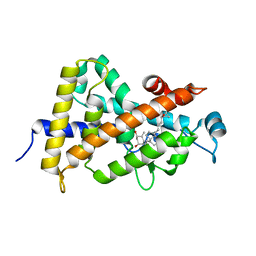 | | Crystal structure of VDR ligand binding domain complexed to calcipotriol. | | Descriptor: | CALCIPOTRIOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
2EVK
 
 | | The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins | | Descriptor: | ACETIC ACID, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qin, J, Perera, R, Lovelace, L.L, Dawson, J.H, Lebioda, L. | | Deposit date: | 2005-10-31 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of thiolate- and carboxylate-ligated ferric H93G myoglobin: models for cytochrome P450 and for oxyanion-bound heme proteins.
Biochemistry, 45, 2006
|
|
2QZA
 
 | | Crystal structure of Salmonella effector protein SopA | | Descriptor: | Secreted effector protein | | Authors: | Diao, J, Chen, J. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
