5MO7
 
 | | Crystal Structure of CK2alpha with N-(3-(((2-chloro-[1,1'-biphenyl]-4-yl)methyl)amino)propyl)methanesulfonamide bound | | Descriptor: | 3-[(3-chloranyl-4-phenyl-phenyl)methylamino]propanamide, Casein kinase II subunit alpha, PHOSPHATE ION | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6T6M
 
 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | Descriptor: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
5MOJ
 
 | | Crystal structure of IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, MacDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
6T4D
 
 | | Crystal structure of Plasmodium falciparum Morn1 | | Descriptor: | Morn1, ZINC ION | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
5MP4
 
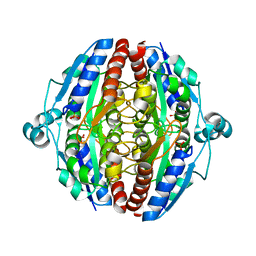 | | The structure of Pst2p from Saccharomyces cerevisiae | | Descriptor: | PHOSPHATE ION, Protoplast secreted protein 2 | | Authors: | Hromic, A, Gruber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure, biochemical and kinetic properties of recombinant Pst2p from Saccharomyces cerevisiae, a FMN-dependent NAD(P)H:quinone oxidoreductase.
Biochim. Biophys. Acta, 1865, 2017
|
|
6T4J
 
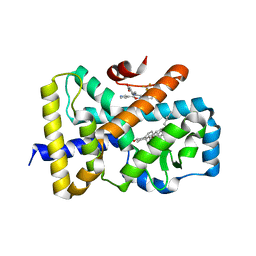 | | ROR(gamma)t ligand binding domain in complex with desmosterol and allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, GLYCEROL, Nuclear receptor ROR-gamma, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5MJH
 
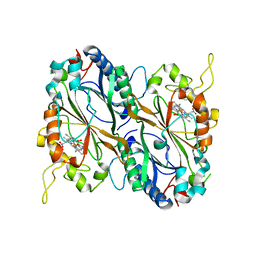 | | X-ray generated oxyferrous/water mixed complex of DtpA from Streptomyces lividans | | Descriptor: | Dye type peroxidase A, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno Chicano, T, Chaplin, A.K, Worrall, J.A.R, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-12-01 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Photoreduction and validation of haem-ligand intermediate states in protein crystals by in situ single-crystal spectroscopy and diffraction.
IUCrJ, 4, 2017
|
|
6T4X
 
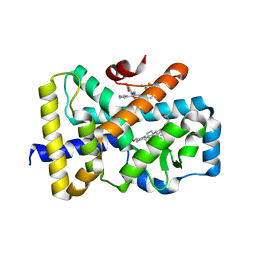 | | ROR(gamma)t ligand binding domain in complex with 25-hydroxycholesterol and allosteric ligand FM26 | | Descriptor: | 25-HYDROXYCHOLESTEROL, 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T5A
 
 | |
5MUX
 
 | |
6T65
 
 | |
5MK8
 
 | |
6T6K
 
 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCINE, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6T82
 
 | |
5MVI
 
 | |
6T8D
 
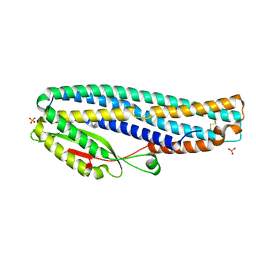 | | The cytotoxin MakB from Vibrio cholerae | | Descriptor: | MakB, SULFATE ION | | Authors: | Persson, K, Nagampalli, R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A tripartite cytolytic toxin formed by Vibrio cholerae proteins with flagellum-facilitated secretion.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5MW8
 
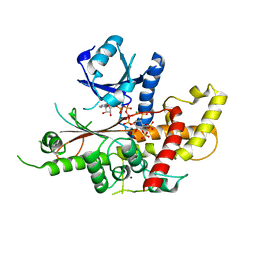 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH ATP and IP5 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
6T6W
 
 | |
6T8T
 
 | |
5MMB
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ434 (compound 6p) | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), GLYCEROL, ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.
J. Med. Chem., 60, 2017
|
|
5MWM
 
 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ZINC ION | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
5MN0
 
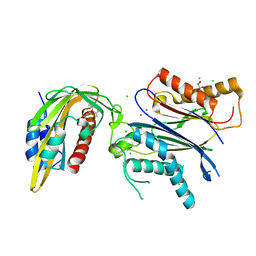 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CHLORIDE ION, CSPYL1, ... | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
6TA8
 
 | |
6TAH
 
 | |
6TBF
 
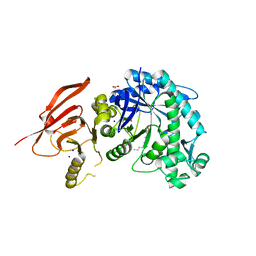 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1~{S},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-5-(octylamino)cyclopentane-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
