2BZ8
 
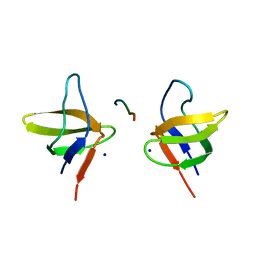 | | N-terminal Sh3 domain of CIN85 bound to Cbl-b peptide | | Descriptor: | SH3-DOMAIN KINASE BINDING PROTEIN 1, SIGNAL TRANSDUCTION PROTEIN CBL-B SH3-BINDING PROTEIN CBL-B, RING FINGER PROTEIN 56, ... | | Authors: | Cardenes, N, Moncalian, G, Bravo, J. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cbl Promotes Clustering of Endocytic Adaptor Proteins
Nat.Struct.Mol.Biol., 12, 2005
|
|
3P4H
 
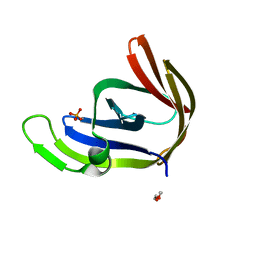 | | Structures of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Zhu, H, Shuman, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
3B8U
 
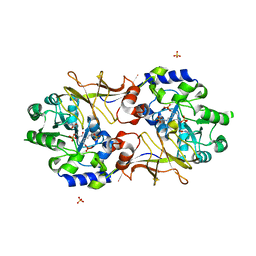 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
7M3Q
 
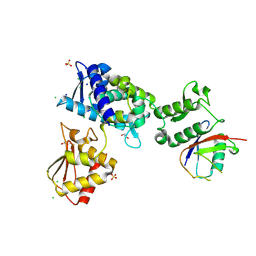 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
1X2W
 
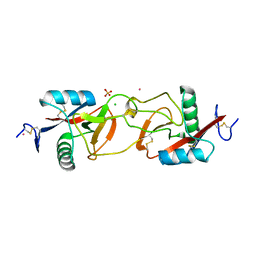 | | Crystal Structure of Apo-Habu IX-bp at pH 4.6 | | Descriptor: | CHLORIDE ION, Coagulation factor IX/X-binding protein A chain, Coagulation factor IX/factor X-binding protein B chain, ... | | Authors: | Suzuki, N, Fujimoto, Z, Morita, T, Fukamizu, A, Mizuno, H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | pH-Dependent Structural Changes at Ca(2+)-binding sites of Coagulation Factor IX-binding Protein
J.Mol.Biol., 353, 2005
|
|
5D70
 
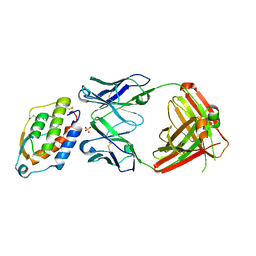 | | Crystal structure of MOR03929, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
1X39
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-3-(ANILINOMETHYL)-5,6,7,8-TETRAHYDRO-5-(HYDROXYMETHYL)-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
3BCO
 
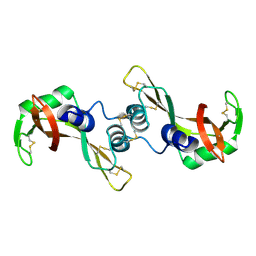 | | Crystal Structure of The Swapped FOrm of P19A/L28Q/N67D BS-RNase | | Descriptor: | Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
1X46
 
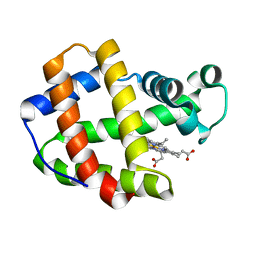 | | Crystal structure of a hemoglobin component (TA-VII) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component VII | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
8RAS
 
 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
7Y4S
 
 | | Structure of human MG53 homo-dimer | | Descriptor: | Tripartite motif-containing protein 72 | | Authors: | Chen, L, Niu, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human MG53 homodimer.
Biochem.J., 479, 2022
|
|
2FUH
 
 | | Solution Structure of the UbcH5c/Ub Non-covalent Complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Brzovic, P.S, Lissounov, A, Hoyt, D.W, Klevit, R.E. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A UbcH5/Ubiquitin Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination.
Mol.Cell, 21, 2006
|
|
1PUB
 
 | |
3B9R
 
 | | SERCA Ca2+-ATPase E2 aluminium fluoride complex without thapsigargin | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Olesen, C, Picard, M, Winther, A.M.L, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of calcium transport by the calcium pump
Nature, 450, 2007
|
|
3H6H
 
 | |
1LJJ
 
 | |
2RKA
 
 | | The Structure of rat cytosolic PEPCK in complex with phosphoglycolate | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
5XXY
 
 | |
1Q03
 
 | | Crystal structure of FGF-1, S50G/V51G mutant | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
8UA2
 
 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (proteolyzed fragment) | | Descriptor: | IODIDE ION, RL2 | | Authors: | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 92, 2024
|
|
8UA5
 
 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (A636-Q776) | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 92, 2024
|
|
1NBX
 
 | | Streptavidin Mutant Y43A at 1.70A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6S9E
 
 | | Tubulin-GDP.AlF complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALUMINUM FLUORIDE, CALCIUM ION, ... | | Authors: | Oliva, M.A, Estevez-Gallego, J, Diaz, J.F, Prota, A.E, Steinmetz, M.O, Balaguer, F.A, Lucena-Agell, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural model for differential cap maturation at growing microtubule ends.
Elife, 9, 2020
|
|
1X38
 
 | | crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
