4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7ZK8
 
 | | ABCB1 L971C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
1V5K
 
 | | Solution structure of the CH domain from mouse EB-1 | | Descriptor: | microtubule-associated protein, RP/EB family, member 1 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from mouse EB-1
To be Published
|
|
1GM8
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1N7J
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
3T2Z
 
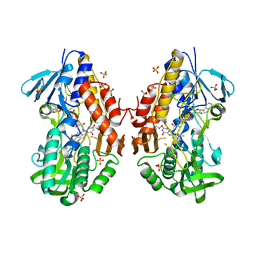 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans | | Descriptor: | 1,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, Solomonson, M, Weiner, J.H, James, M.N. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2994 Å) | | Cite: | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification.
J.Mol.Biol., 398, 2010
|
|
4DAF
 
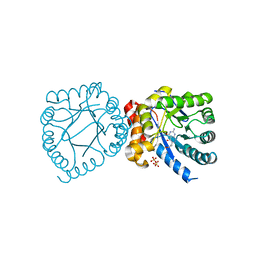 | | Crystal structure of B. anthracis DHPS with compound 19 | | Descriptor: | (2R)-2-(7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)propanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
4MBG
 
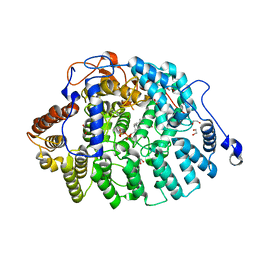 | | Crystal structure of Aspergillus fumigatus protein farnesyltransferase binary complex with farnesyldiphosphate | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
2ZB0
 
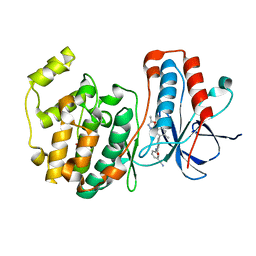 | | Crystal structure of P38 in complex with biphenyl amide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(3-cyanophenyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)-4-biphenylcarboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4M8X
 
 | | GS-8374, a Novel Phosphonate-Containing Inhibitor of HIV-1 Protease, Effectively Inhibits HIV PR Mutants with Amino Acid Insertions | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Grantz saskova, K, Brynda, J, Rezacova, P, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-08-14 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GS-8374, a prototype phosphonate-containing inhibitor of HIV-1 protease, effectively inhibits protease mutants with amino acid insertions.
J.Virol., 88, 2014
|
|
1JNV
 
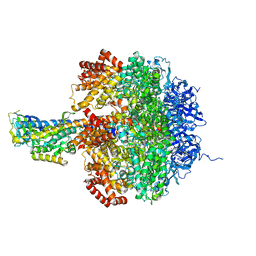 | | The Conformation of the Epsilon and Gamma Subunits within the E. coli F1 ATPase | | Descriptor: | ATP SYNTHASE ALPHA CHAIN, ATP SYNTHASE BETA CHAIN, ATP SYNTHASE EPSILON CHAIN, ... | | Authors: | Hausrath, A.C, Capaldi, R.A, Matthews, B.W. | | Deposit date: | 2001-07-25 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | The conformation of the epsilon- and gamma-subunits within the Escherichia coli F(1) ATPase.
J.Biol.Chem., 276, 2001
|
|
3VS7
 
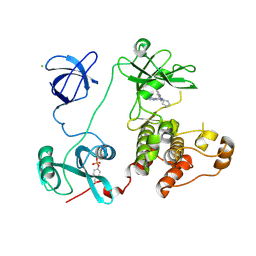 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Honda, K, Niwa, H, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
5CH7
 
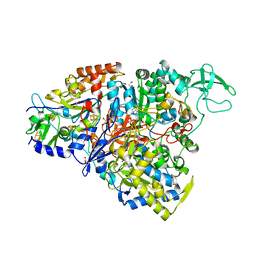 | | Crystal structure of the perchlorate reductase PcrAB - Phe164 gate switch intermediate - from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
6SQC
 
 | | Crystal structure of complex between nuclear coactivator binding domain of CBP and [1040-1086]ACTR containing alpha-methylated Leu1055 and Leu1076 | | Descriptor: | 1,2-ETHANEDIOL, Maltose/maltodextrin-binding periplasmic protein,CREB-binding protein, Nuclear receptor coactivator 3, ... | | Authors: | Bauer, V, Schmidtgall, B, Gogl, G, Dolenc, j, Osz, J, Kostmann, C, Mitschler, A, Cousido-Siah, A, Rochel, N, Trave, G, Kieffer, B, Torbeev, V. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational editing of intrinsically disordered protein by alpha-methylation.
Chem Sci, 12, 2020
|
|
4QII
 
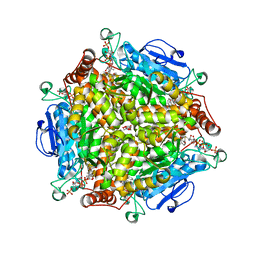 | | Crystal Structure of type II MenB from Mycobacteria tuberculosis | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, Salicylyl CoA, TRIETHYLENE GLYCOL | | Authors: | Song, H.G, Tse, Y.S, Sung, H.P, Guo, Z.H. | | Deposit date: | 2014-05-31 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-dependent active-site closure revealed in the crystal structure of Mycobacterium tuberculosis MenB complexed with product analogues
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7JVN
 
 | |
3VRZ
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
4C4Z
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A8 | | Descriptor: | 1-ethyl-3-naphthalen-1-ylurea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2VC9
 
 | | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5VB2
 
 | |
5C38
 
 | | Crystal structure of AABB + UDP-C-Gal + DI | | Descriptor: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Gagnon, S, Meloncelli, P, Zheng, R.B, Haji-Ghassemi, O, Johal, A.R, Borisova, S, Lowary, T.L, Evans, S.V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Structures of the Human ABO(H) Blood Group Enzymes in Complex with Donor Analogs Reveal That the Enzymes Utilize Multiple Donor Conformations to Bind Substrates in a Stepwise Manner.
J.Biol.Chem., 290, 2015
|
|
